Popular information
Popular science background:
DNA repair – providing chemical stability for life (pdf)
Populärvetenskaplig information:
DNA-reparation – en grund för livets kemiska stabilitet (pdf)

The Nobel Prize in Chemistry 2015
From one cell to another, from one generation to the next. The genetic information that governs how human beings are shaped has flowed through our bodies for hundreds of thousands of years. It is constantly subjected to assaults from the environment, yet it remains surprisingly intact. Tomas Lindahl, Paul Modrich and Aziz Sancar are awarded the Nobel Prize in Chemistry 2015 for having mapped and explained how the cell repairs its DNA and safeguards the genetic information.
DNA repair – providing chemical stability for life
The foundation of who you are was created when 23 chromosomes from a sperm combined with 23 chromosomes from an egg. Together, they formed the original version of your genome, your genetic material. All the genetic information required to create you was present in that fusion. If someone had pulled out the DNA molecules from this first cell and laid them in a row, it would have been two metres long.
When the fertilised egg subsequently divided, the DNA molecules were copied and the daughter cell also obtained a full set of chromosomes. After that, the cells divided again; two became four, four became eight. After the first week you consisted of 128 cells, each one with its own set of genetic material. The total length of your DNA began to approach 300 metres.
Today – many, many billions of cell divisions later – your DNA could stretch all the way to the sun and back, around 250 times. Even though your genetic material has been copied so many times, the most recent copy is remarkably similar to the original that was once created in the fertilised egg. This is where life’s molecules display their greatness, because from a chemical perspective this ought to be impossible. All chemical processes are prone to random errors. Additionally, your DNA is subjected on a daily basis to damaging radiation and reactive molecules. In fact, you ought to have been a chemical chaos long before you even developed into a foetus.
Your DNA is monitored by a swarm of proteins
Our DNA remains astonishingly intact, year after year, due to a host of molecular repair mechanisms: a swarm of proteins that monitor the genes. They continually proof-read the genome and repair any damage that has occurred. The Nobel Prize in Chemistry 2015 is awarded to Tomas Lindahl, Paul Modrich and Aziz Sancar for having mapped these fundamental processes at the molecular level. Their systematic work has made a decisive contribution to the understanding of how the living cell functions, as well as providing knowledge about the molecular causes of several hereditary diseases and about mechanisms behind both cancer development and aging.
Tomas Lindahl, Paul Modrich and Aziz Sancar have, independently of each other, mapped several processes for DNA repair that are relevant to humans. The story begins with Tomas Lindahl, born in the same country as Alfred Nobel.
Life exists – so DNA must be repairable
“How stable is DNA, really?”, Tomas Lindahl started wondering towards the end of the 1960s. At the time, the scientific community believed that the DNA molecule – the foundation of all life – was extremely resilient; anything else was simply out of the question. Evolution does require mutations, but only a limited number per generation. If genetic information were too unstable no multi-cellular organisms would exist. During his postdoc at Princeton University, USA, Tomas Lindahl worked on the RNA molecule, a molecular cousin to DNA. It did not go well. In his experiment he had to heat RNA, but this inevitably led to the molecules’ rapid degradation. It was well known that RNA was more sensitive than DNA, but if RNA was destroyed so quickly when subjected to heat, could DNA molecules really be stable for a lifetime? This question took hold in Lindahl’s mind.
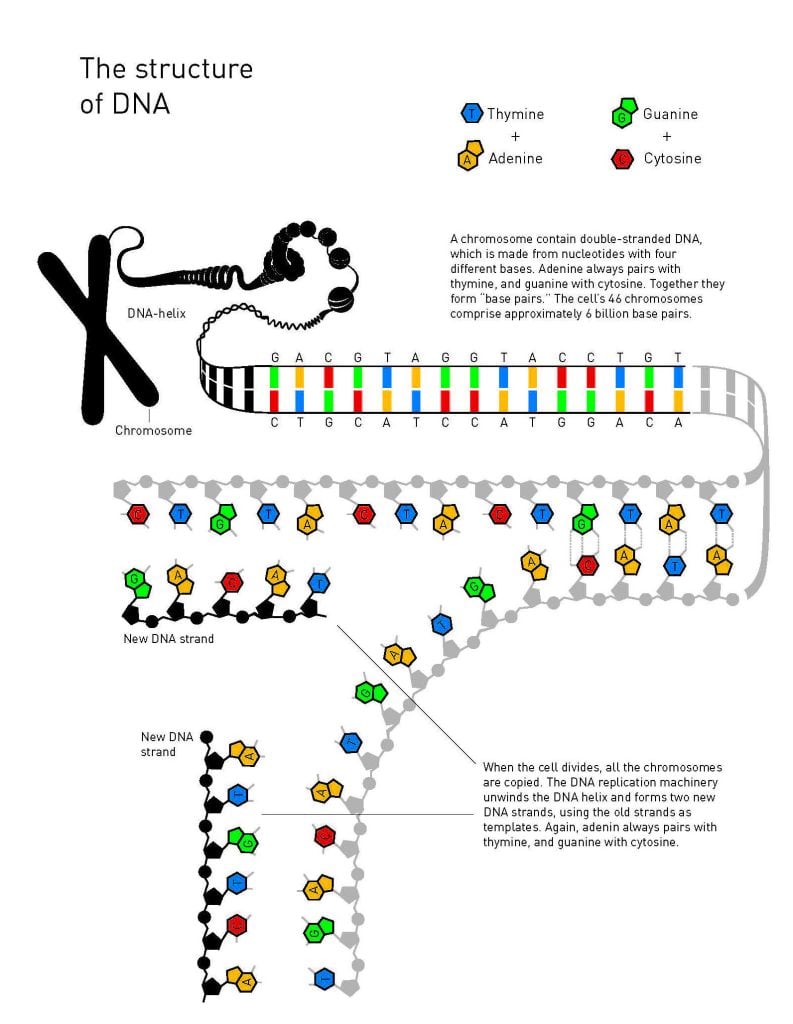
It would be a few years before he began to look for an answer to that question, and by then he had moved back to Sweden and Karolinska Institutet in Stockholm. Some straightforward experiments proved that his suspicions were correct: DNA underwent a slow but noticeable decay. Lindahl estimated that there were thousands of potentially devastating injuries to the genome every day, a frequency that was clearly incompatible with human existence on Earth. His conclusion was that there must be molecular systems for repairing all these DNA defects and, with this idea, Tomas Lindahl opened the door on an entirely new field of research.
Special enzymes remove damage in DNA
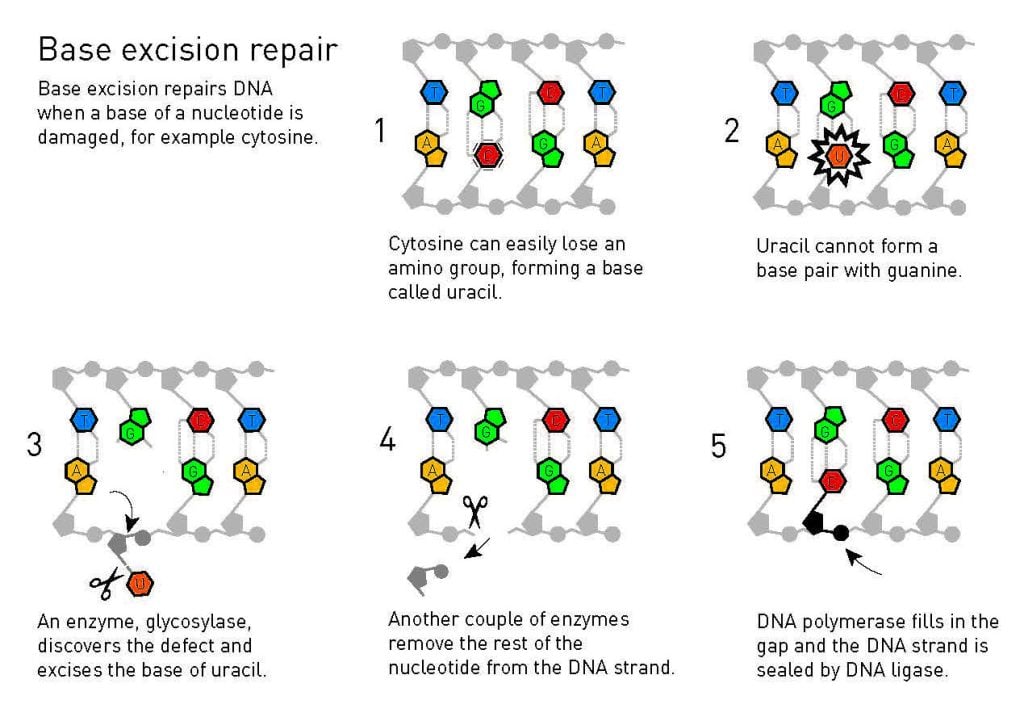
Using bacterial DNA which, just like human DNA, consists of nucleotides with the bases adenine, guanine, cytosine, and thymine, Tomas Lindahl began to look for repair enzymes. One chemical weakness in DNA is that cytosine easily loses an amino group, which can lead to the alteration of genetic information. In DNA’s double helix, cytosine always pairs with guanine, but when the amino group disappears, the damaged remains tend to pair with adenine. Therefore, if this defect is allowed to persist, a mutation will occur the next time DNA is replicated. Lindahl realised that the cell must have some protection against this, and was able to identify a bacterial enzyme that removes damaged remains of cytosines from DNA. In 1974, he published his findings.
Tomas Lindahl puts together the pieces of base excision repair
This was the start of 35 years of successful work, during which Tomas Lindahl has found and examined many of the proteins in the cell’s toolbox for DNA repair. In the beginning of the 1980s, a relationship took him to Great Britain, where he took up a position at the Imperial Cancer Research Fund in London. In 1986, he became director of the newly founded Clare Hall Laboratory, subsequently known for its scientific creativity.
Bit by bit, Lindahl pieced together a molecular image of how base excision repair functions, a process in which glycosylases, enzymes similar to the one he had found in 1974, are the first step in the DNA repair process. Base excision repair also occurs in human beings and, in 1996, Tomas Lindahl managed to recreate the human repair process in vitro.
The decisive factor for Tomas Lindahl was the realisation that DNA inevitably undergoes change, even when the molecule is located in the cell’s protective environment. However, it had long been known that DNA can be damaged by environmental assaults such as UV radiation. The mechanism used by the majority of cells to repair UV damage, nucleotide excision repair, was mapped by Aziz Sancar, born in Savur, Turkey, and professionally active in the USA.
Biochemistry preferable to life as a doctor
Aziz Sancar’s fascination with life’s molecules developed while he was studying for a medical degree in Istanbul. After graduating, he worked for a few years as phycisian in the Turkish countryside, but in 1973 he decided to study biochemistry. His interest was piqued by one phenomenon in particular: when bacteria are exposed to deadly doses of UV radiation, they can suddenly recover if they are illuminated with visible blue light. Sancar was curious about this almost magical effect; how did it function chemically?
Claud Rupert, an American, had studied this phenomenon and Aziz Sancar joined his laboratory at the University of Texas in Dallas, USA. In 1976, using that time’s blunt tools for molecular biology, he succeeded in cloning the gene for the enzyme that repairs UV-damaged DNA, photolyase, and also in getting bacteria to over-produce the enzyme. This work became a doctoral dissertation, but people were hardly impressed; three applications for postdoc positions resulted in as many rejections. His studies of photolyase had to be shelved. In order to continue working on DNA repair, Aziz Sancar took up a position as laboratory technician at the Yale University School of Medicine, a leading institution in the field. Here he started the work that would eventually result in the Nobel Prize in Chemistry.
Aziz Sancar – investigating how cells repair UV damage
By then it was clear that bacteria have two systems for repairing UV damage: in addition to light-dependent photolyase, a second system that functions in the dark had been discovered. Aziz Sancar’s new colleagues at Yale had studied this dark system since the mid-1960s, using three UV-sensitive strains of bacteria that carried three different genetic mutations: uvrA, uvrB and uvrC.
As in his previous studies of photolyase, Sancar began investigating the molecular machinery of the dark system. Within a few years he had managed to identify, isolate and characterise the enzymes coded by the genes uvrA, uvrB and uvrC. In ground-breaking in vitro experiments he showed that these enzymes can identify a UV-damage, then making two incisions in the DNA strand, one on each side of the damaged part. A fragment of 12-13 nucleotides, including the injury, is then removed.
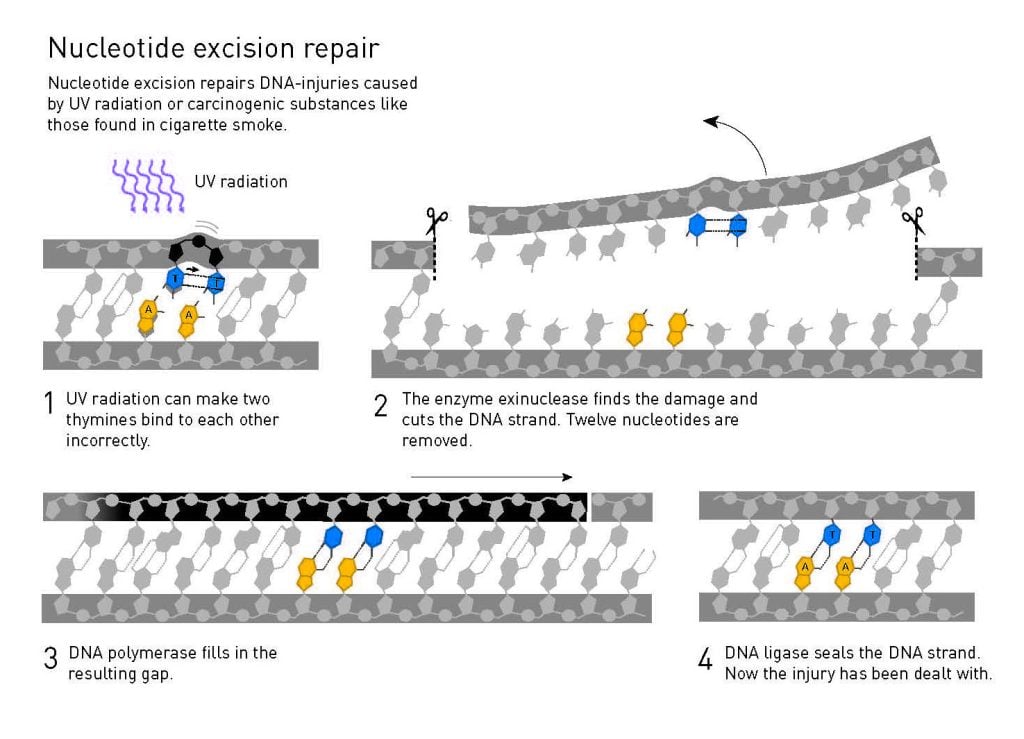
Similar mechanisms for UV damage repair in humans and bacteria
Aziz Sancar’s ability to generate knowledge about the molecular details of the process changed the entire research field. He published his findings in 1983. His achievements led to an offer of an associate professorship in biochemistry at the University of North Carolina at Chapel Hill. There, and with the same precision, he mapped the next stages of nucleotide excision repair. In parallel with other researchers, including Tomas Lindahl, Sancar investigated nucleotide excision repair in humans. The molecular machinery that excises UV damage from human DNA is more complex than its bacterial counterpart but, in chemical terms, nucleotide excision repair functions similarly in all organisms.
So, what happened to Sancar’s initial interest in photolyase? Well, he eventually returned to this enzyme, uncovering the mechanism responsible for reviving the bacteria. In addition, he helped to demonstrate that a human equivalent to photolyase helps us set the circadian clock.
Time to turn to the work of Paul Modrich. He also began with a vague idea about a repair mechanism, which he then chiselled out in elegant molecular detail.
It pays off to learn about “DNA stuff”
Paul Modrich grew up in a small town in northern New Mexico, USA. The diversity of the expansive landscape spurred his interest in nature, but one day his father, a biology teacher, said: “You should learn about this DNA stuff.” This was in 1963, the year after James Watson and Francis Crick had been awarded the Nobel Prize for discovering the structure of DNA.
A few years later, that “DNA stuff” really became central to Paul Modrich’s life. Early in his research career, as doctoral student at Stanford, during his postdoc at Harvard, and as an assistant professor at Duke University, he examined a series of enzymes that affect DNA: DNA ligase, DNA polymerase and the restriction enzyme EcoRI. When he subsequently, towards the end of the 1970s, shifted his attention to the enzyme Dammethylasehe stumbled over another piece of “DNA stuff” that would come to occupy him for a large part of his scientific career.
Interweaving two strands of research
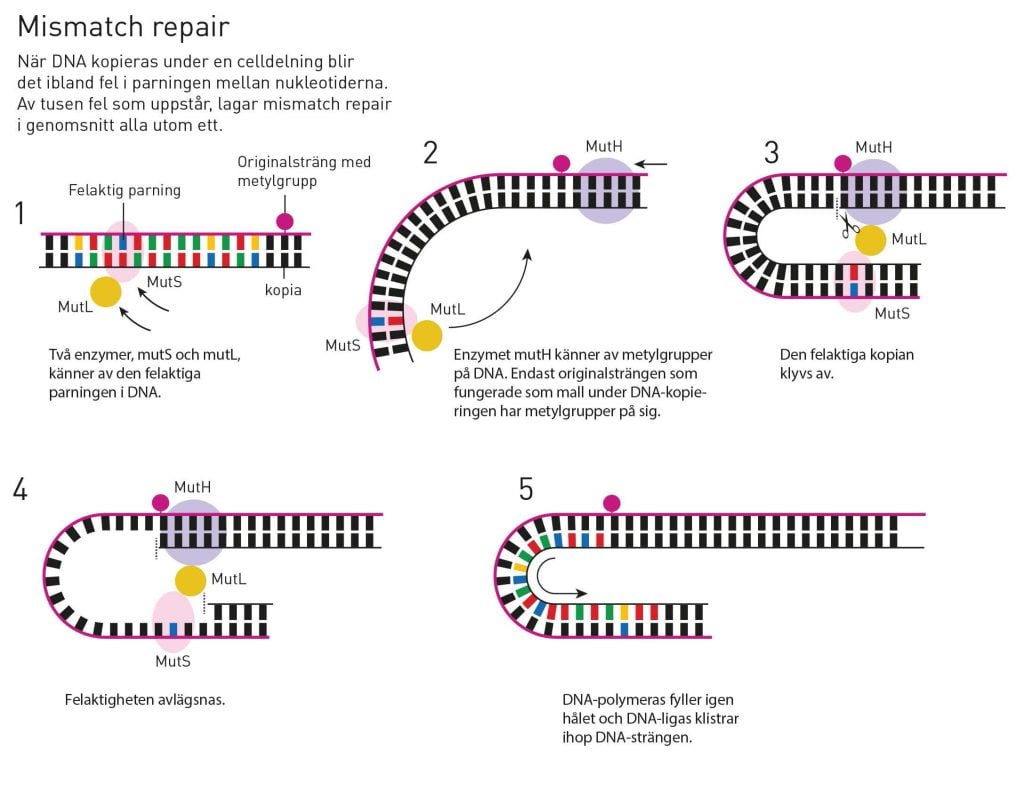
Dam methylase couples methyl groups to DNA. Paul Modrich showed that these methyl groups could function as signposts, helping a particular restriction enzyme to cut the DNA strand at the correct location. However, only a few years earlier, Matthew Meselson, a molecular biologist at Harvard University, had suggested a different signalling function for the methyl groups on DNA.
Using some molecular biology artistry, Meselson had constructed a bacterial virus with several occurrences of mismatching bases in the DNA. For instance, A could be placed opposite C, instead of T. When he let these viruses infect bacteria, the bacteria corrected the mismatches. No one knew why the bacteria had developed this function, but in 1976 Meselson speculated, among other things, that it could be a repair mechanism that corrected the faulty matches that sometimes occur when DNA is replicated. If that was the case, Meselson continued, perhaps the methyl groups on the DNA helped the bacteria identify which strand to use as template during correction. As the new DNA strand, the faulty replica, was still unmethylated, maybe that was how it could be identified and corrected?
Here – in the methylation of DNA – Paul Modrich’s and Matthew Meselson’s paths crossed. Working together, they created a virus with a number of mismatches in its DNA. This time, Modrich’s dam methylase was also used to add methyl groups to one of the DNA strands. When these viruses infected bacteria, the bacteria consistently corrected the DNA strand that lacked methyl groups. Modrich and Meselson’s conclusion was that DNA mismatch repair is a natural process that corrects mismatches that occur when DNA is copied, recognising the defect strand by its unmethylated state.
Paul Modrich – illustrating DNA mismatch repair
For Paul Modrich, this discovery kick-started a decade of systematic work, cloning and mapping one enzyme after the other in the mismatch repair process. Towards the end of the 1980s, he was able to recreate the complex molecular repair mechanism in vitro and study it in great detail. This work was published in 1989.
Paul Modrich, just like Tomas Lindahl and Aziz Sancar, has also studied the human version of the repair system. Today we know that all but one out of a thousand errors that occur when the human genome is copied, are corrected by mismatch repair. However, in human mismatch repair, we still do not know for sure how the original strand is identified. DNA methylation has other functions in our genome to that of bacteria, so something else must govern which strand gets corrected – and exactly what remains to be clarified.
Defects in the repair systems cause cancer
Besides base excision repair, nucleotide excision repair, and mismatch repair, there are several other mechanisms that maintain our DNA. Every day, they fix thousands of occurrences of DNA damage caused by the sun, cigarette smoke or other genotoxic substances; they continuously counteract spontaneous alterations to DNA and, for each cell division, mismatch repair corrects some thousand mismatches. Our genome would collapse without these repair mechanisms. If just one component fails, the genetic information changes rapidly and the risk of cancer increases. Congenital damage to the nucleotide excision repair process causes the disease xeroderma pigmentosum; individuals who suffer from this disease are extremely sensitive to UV radiation and develop skin cancer after exposure to the sun. Defects in DNA mismatch repair increase the risk of developing hereditary colon cancer, for instance.
In fact, in many forms of cancer, one or more of these repair systems have been entirely or partially switched off. This makes the cancer cells’ DNA unstable, which is one reason why cancer cells often mutate and become resistant to chemotherapy. At the same time, these sick cells are even more dependent on the repair systems that are still functioning; without these, their DNA will become too damaged and the cells will die. Researchers are attempting to utilise this weakness in the development of new cancer drugs. Inhibiting a remaining repair system allows them to slow down or completely stop the growth of the cancer. One example of a pharmaceutical that inhibits a repair system in cancer cells is olaparib.
In conclusion, the basic research carried out by the 2015 Nobel Laureates in Chemistry has not only deepened our knowledge of how we function, but could also lead to the development of lifesaving treatments. Or, in the words of Paul Modrich: “That is why curiosity-based research is so important. You never know where it is going to lead … A little luck helps, too.”
Links and further reading
Popular science articles
Howard Hughes Medical Institute, Biography Paul Modrich.
http://www.hhmi.org/scientists/paul-l-modrich
Weston, K. (2014) Country Life: Repair and Replication. In Blue Skies and Bench Space: Adventures in Cancer Research. Long Island, New York, Cold Spring Harbor Laboratory Press.
http://blueskiesbenchspace.org/index.php?pag=4
Zagorski, N. (2005) Profile of Aziz Sancar, Proc. Nat. Acad. Sci. USA, 102(45), 16125–16127.
http://www.pnas.org/content/102/45/16125.full.pdf
Videos
Howard Hughes Medical Institute (2003) Mismatch repair.
http://www.hhmi.org/biointeractive/mismatch-repair
Interview with T. Lindahl (2015) Cancer Research UK.
https://www.youtube.com/watch?v=FHlnqiEQig0&index=16&list=PL_bJU93S6g0sCs1CQ_eh2o_z7ztU-QkE
Scientific articles
Lahue, R. S, Au, K. G. and Modrich, P. (1989) DNA Mismatch Correction in a Defined System, Science, 245(4914), 160–164.
Lindahl, T. (1974) An N-Glycosidase from Escherichia coli That Releases Free Uracil from DNA Containing Deaminated Cystosine Residues, Proc. Nat. Acad. Sci. USA, 71(9), 3649–3653.
Sancar, A. and Rupp, W. D. (1983) A Novel Repair Enzyme: UVRABC Excision Nuclease of Escherichia coli
Cuts a DNA Strand on Both Sides of the Damaged Region, Cell, 33(1), 249–260.
The Laureates
TOMAS LINDAHL
Swedish citizen. Born 1938 in Stockholm, Sweden. Ph.D. 1967 from Karolinska Institutet, Stockholm, Sweden. Professor of Medical and Physiological Chemistry at University of Gothenburg 1978–82.
Emeritus group leader at Francis Crick Institute and Emeritus director of Cancer Research UK at Clare Hall Laboratory, Hertfordshire, UK.
http://crick.ac.uk/research/a-zresearchers/emeritus-scientists/tomas-lindahl/
PAUL MODRICH
U.S. citizen. Born 1946. Ph.D. 1973 from Stanford University, Stanford, CA, USA. Investigator at Howard Hughes Medical Institute and James B. Duke Professor of Biochemistry at Duke University School of Medicine, Durham, NC, USA.
http://www.biochem.duke.edu/paul-lmodrich-primary
AZIZ SANCAR
U.S. and Turkish citizen. Born 1946 in Savur, Turkey. Ph.D. 1977 from University of Texas, Dallas, TX, USA. Sarah Graham Kenan Professor of Biochemistry and Biophysics, University of North Carolina School of Medicine, Chapel Hill, NC, USA.
http://www.med.unc.edu/biochem/people/faculty/primary/asancar
Science Editors: Claes Gustafsson, Gunnar von Heijne and Sara Snogerup Linse, the Nobel Committee for Chemistry
Text: Ann Fernholm
Translation: Peter Wennersten
Illustrations: © Johan Jarnestad/The Royal Swedish Academy of Sciences
Editor: Carl-Victor Heinold
© The Royal Swedish Academy of Sciences
Nobel Prizes and laureates
Six prizes were awarded for achievements that have conferred the greatest benefit to humankind. The 12 laureates' work and discoveries range from proteins' structures and machine learning to fighting for a world free of nuclear weapons.
See them all presented here.
