Popular information
Information for the public (pdf)
Populärvetenskaplig information (pdf)
The Nobel Prize in Physiology or Medicine 2006
jointly to
Andrew Fire and Craig Mello
“for their discovery of RNA interference – gene silencing by double-stranded RNA”
The Nobel Laureates in Physiology or Medicine for 2006, the American scientists Andrew Fire and Craig Mello, have discovered a fundamental mechanism of gene regulation. They have shown that double-stranded RNA molecules can silence a gene by elimination of the mRNA corresponding to that gene. This is called RNA interference (RNAi) and occurs in both plants and animals.
How proteins are formed
An adult human being has about 100 000 billion cells. The nucleus of each cell carries all our genetic material: a total of two meters of DNA containing some 30 000 genes. When genes are expressed (activated), genetic information is copied from DNA to messenger molecules, messenger RNA (mRNA), which then orchestrate formation of proteins. Proteins are involved in the life processes of all organisms.
Expression of genes is directed by the cellular machinery that copies (transcribes) DNA into mRNA. The human body contains a few hundred different types of cells. Formation and renewal of these cells is regulated through gene activity.
All genes are not expressed in each cell: different sets of genes are active in different types of cells. Nerve cells express genes different from those expressed by muscle cells or liver cells. Which genes are active at any given point in time is regulated by transcription factors, proteins that govern how genes are read.
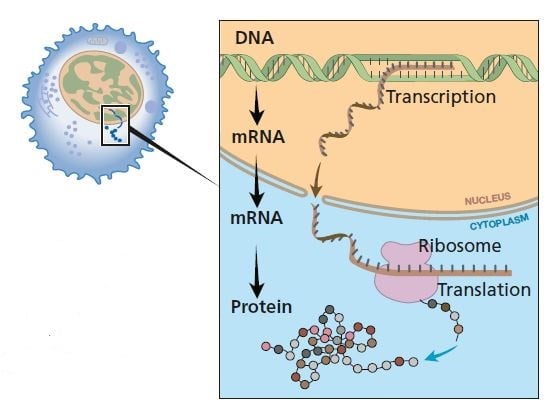
When a protein is to be formed, single-stranded messenger RNA is made as a copy of one of the strands of DNA (transcription). mRNA moves from the nucleus to the cytoplasm where the information is decoded (translation). Proteins are formed in ribosomes.
© The Nobel Assembly at Karolinska Institutet
© The Nobel Assembly at Karolinska Institutet
How does RNA interference work?
Andrew Fire and Craig Mello published their discovery of RNA interference in 1998. Their surprising discovery was that double-stranded RNA (dsRNA) plays an active part in regulating gene activity. These RNA molecules control degradation of mRNA molecules by the biochemical machinery. In this way, the expression of the gene that encodes (gives rise to) a specific protein can be abolished: the gene is silenced. The result is that production of the protein is turned off.
RNA interference is a natural mechanism of great importance in regulation of the activity of our genes. It has also become a highly useful research tool. In basic research, RNA interference is primarily used to study the function of specific genes. The hope is that, in the future, RNA interference may be used to treat diseases.
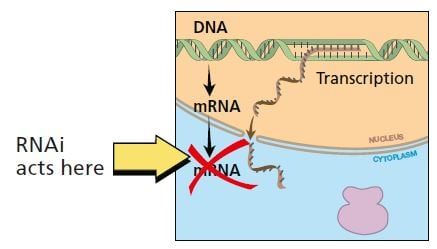
In RNA interference, RNA in doublestranded form breaks down the mRNA for a specific gene, thus stopping production of protein.
© The Nobel Assembly at Karolinska Institutet
© The Nobel Assembly at Karolinska Institutet
Puzzling antisense and plant experiments
RNA molecules were long believed to serve only as messengers, bearing genetic information from DNA. But in the early 1980s it was discovered that single-stranded RNA molecules in the bacterium Escherichia coli can bind to mRNA. This inactivates mRNA and therefore prevents it from passing on the genetic information. As a result, the protein encoded by the mRNA is no longer formed. Similar “antisense” molecules were later discovered in the nematode Caenorhabditis elegans.
These findings inspired hope that antisense RNA could be used as a form of gene therapy. Many saw use of “tailor-made” antisense molecules to block the production of faulty proteins as a potential new method of treating hereditary diseases and cancer.
Antisense techniques turned out to work well in plants. For example, a tomato strain genetically modified to ripen more slowly than usual has been created in this manner. But attempts to use antisense RNA experimentally in animals have not been equally successful. Sometimes the method has worked, sometimes it has not, and it turned out to be difficult to get consistent results when experiments were repeated.

Plant scientists wanted to make petunias even more beautiful by enhancing the red of the petals. But sometimes the flower lost all its color!
© The Nobel Assembly at Karolinska Institutet
© The Nobel Assembly at Karolinska Institutet
Around 1990, a series of observations were obtained that were difficult to explain. The most puzzling findings were reported by plant scientists who were trying to intensify the hue of red petunias. They inserted a gene that controls formation of red pigment – but in some cases the result was that the flowers lost their color altogether! It turned out that the mRNA for the pigment had disappeared. The cause of these unexpected effects remained enigmatic until the discoveries of Andrew Fire and Craig Mello solved the riddle.
The key experiment
The crucial experiment that led to the discovery of RNA interference was carried out with the nematode (roundworm) Caenorhabditis elegans, a millimeter-long animal that has played an important role in many biomedical discoveries.
Caenorhabditis elegans
© The Nobel Assembly at Karolinska Institutet
© The Nobel Assembly at Karolinska Institutet
In the 1990s, Andrew Fire and Craig Mello were studying how gene expression is regulated in C. elegans. Among other things, they were examining mRNA that encodes a protein involved in the nematode’s ability to move. They injected the nematode gonads with sense RNA (a molecule identical to a segment of mRNA). This had no visible effect on the nematodes or on their offspring. Fire and Mello injected other nematodes with antisense RNA that could bind to the corresponding mRNA for the muscle protein, and again nothing happened. But when they injected a mixture of sense RNA and antisense RNA, the nematode offspring moved in an odd, twitching fashion. Similar movement patterns could be seen in nematodes with a defective muscle protein gene. What was the explanation for this surprising finding?
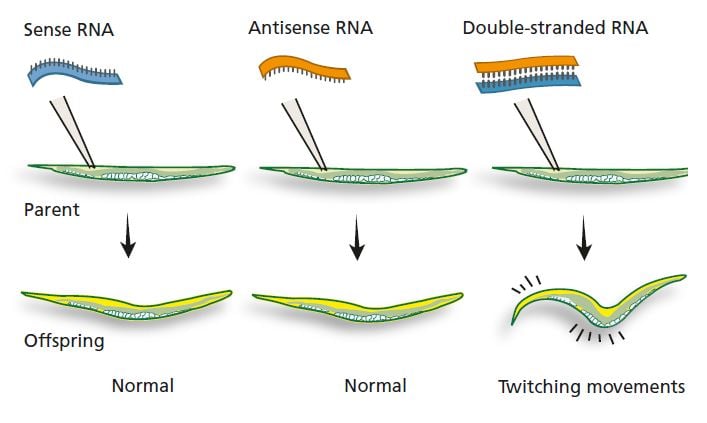
Neither injection of sense RNA for the muscle protein nor of the corresponding antisense RNA had any effect. But after simultaneous injection of both sense and antisense RNA (which bind to each other and form double-stranded RNA) the nematode offspring showed the same twitching movements as nematodes whose muscle protein gene was defective.
© The Nobel Assembly at Karolinska Institutet
© The Nobel Assembly at Karolinska Institutet
Genes are silenced when mRNA is eliminated
Together, sense RNA and the corresponding antisense RNA make up double-stranded RNA, and it seemed as if this could silence the gene that encoded the muscle protein. But how? After all, double-stranded RNA molecules have no free chains that can bind and inactivate mRNA molecules. One would have expected that only single-stranded antisense RNA would bind to the corresponding mRNA and silence the gene.
Andrew Fire and Craig Mello pursued the problem by injecting double-stranded RNA carrying the genetic code of several other C. elegans genes. With a special staining technique they could show mRNA for genes that were active in nematode embryos. When the gene was active, the embryo could be stained because its cells contained the mRNA. Antisense RNA binding to the mRNA for this gene reduced the staining somewhat. But doublestranded RNA (sense RNA plus antisense RNA) eliminated the staining entirely – the mRNA had disappeared and the gene had been silenced.
In all the experiments Fire and Mello did with nematode genes, double-stranded RNA extinguished the gene it had been patterned on. This stopped the formation of the protein encoded by the gene. Double-stranded RNA could interfere with the expression of genes; thus the phenomenon was called RNA interference.

The activity of a gene in the embryo of the nematode C. elegans can be studied in the laboratory. When the gene is expressed, the nematode embryo can be stained. When Fire and Mello injected antisense RNA for the gene, the staining was slightly less intense. But when they injected the corresponding double-stranded RNA (sense RNA plus antisense RNA) the staining disappeared entirely.
© The Nobel Assembly at Karolinska Institutet. Photos from Nature, Feb. 1998
© The Nobel Assembly at Karolinska Institutet
Fire’s and Mello’s conclusions
In their article in Nature, 19 February 1998, Andrew Fire and Craig Mello drew a series of conclusions from their simple but elegant experiments with C. elegans genes:
- Gene silencing was highly effective when double-stranded RNA was injected but weak or non-existent after injection of single-stranded RNA (sense RNA or antisense RNA for the gene).
- The mRNA affected by the double-stranded RNA disappeared – it was apparently broken down and eliminated.
- The double-stranded RNA injected must match the mature, “trimmed” mRNA sequence for the Interference could not be elicited by intron sequences (that is, segments of molecules that do not contain coding information). This implies that the interference takes place after transcription, probably in the cytoplasm rather than in the cell nucleus.
- Only the mRNA corresponding to the sense RNA strand of the double-stranded RNA was silenced – no other mRNA in the cells was affected. RNA interference was specific for the gene with a code corresponding to that of the mRNA molecule.
- Just a few double-stranded RNA molecules were needed to silence a gene completely. The effect was so strong that Fire and Mello suggested that enzymes were involved in the process.
- The effect of double-stranded RNA could spread from cell to cell and from tissue to tissue and could even be passed on to offspring.
Explained previous results
The findings of Andrew Fire and Craig Mello explained some results that had until then been perplexing, including the peculiar experiment where red petunias lost their color when they received a gene intended to make them even more red. At the end of the article in Nature, Fire and Mello speculated that organisms might use double-stranded RNA naturally, as a way of silencing genes.
Fire’s and Mello’s discoveries opened up a completely new research field. Over the next few years, many components of the RNA interference machinery were identified. Interference is an intricate process where a multitude of proteins and protein complexes interact. The mechanism differs considerably from the simple antisense mechanism discovered in bacteria in the early 1980s.
Still, in simplified terms, RNA interference is ultimately a type of antisense mechanism that results in mRNA being broken down and eliminated. The corresponding gene is thus silenced and cannot form the protein it encodes. RNA interference has been shown to occur in essentially all organisms whose cells contain a nucleus.
Regulation of gene expression
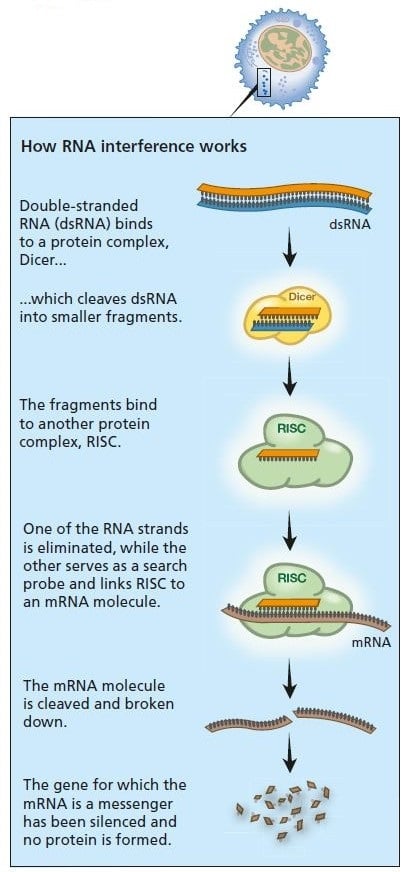
What are the natural roles of RNA interference in organisms? We now know that several hundred genes in our genome encode small RNA molecules collectively called microRNA, which contain segments of codes from other genes. Precursors of these microRNA molecules form hairpin structures of double-stranded RNA (see figure below). It is these molecules that activate the RNA interference machinery. The result is that mRNA from a gene with a similar coding sequence is broken down or blocked so that the protein cannot be produced.
Use of microRNA molecules to regulate gene expression has turned out to be fundamental for eukaryote cells – from single-cell organisms to humans. RNA interference plays an important role in switching off genes during an organism’s development and to control cellular function.
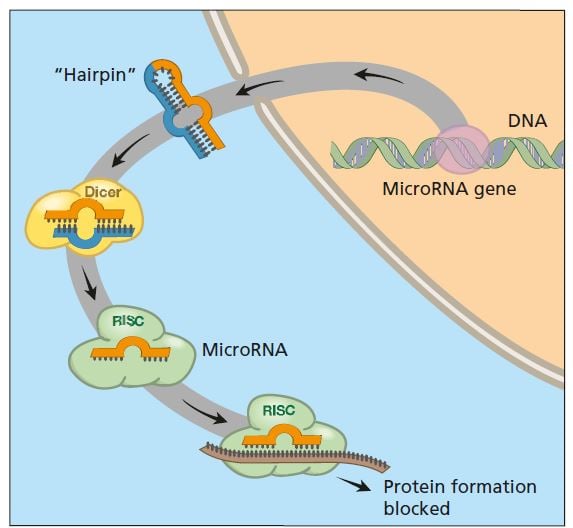
We have hundreds of different genes that encode small RNA (collectively, microRNA) whose precursors can form double-stranded RNA. These can activate the RNA interference process and thus switch off the activity of various genes with matching segments.
© The Nobel Assembly at Karolinska Institutet
© The Nobel Assembly at Karolinska Institutet
Defence against viruses
Humans and other vertebrates have advanced immune systems as defence against invading micro-organisms. Interferons and various types of T cells protect against viral infections.
But among more primitive organisms that lack an efficient immune system, RNA interference has proved to be an important mechanism for protection against viral infections. Some viruses have genetic material that consists of double-stranded RNA, and many viruses with single-stranded RNA have their RNA in double-stranded form at some point during the reproductive life cycle within the host cell.
A virus infection starts when viral RNA enters the cell (see figure below). Double-stranded RNA binds to Dicer and is broken down into smaller fragments. This in turn activates the RISC complex: viral RNA is broken down and the cell survives the infection.
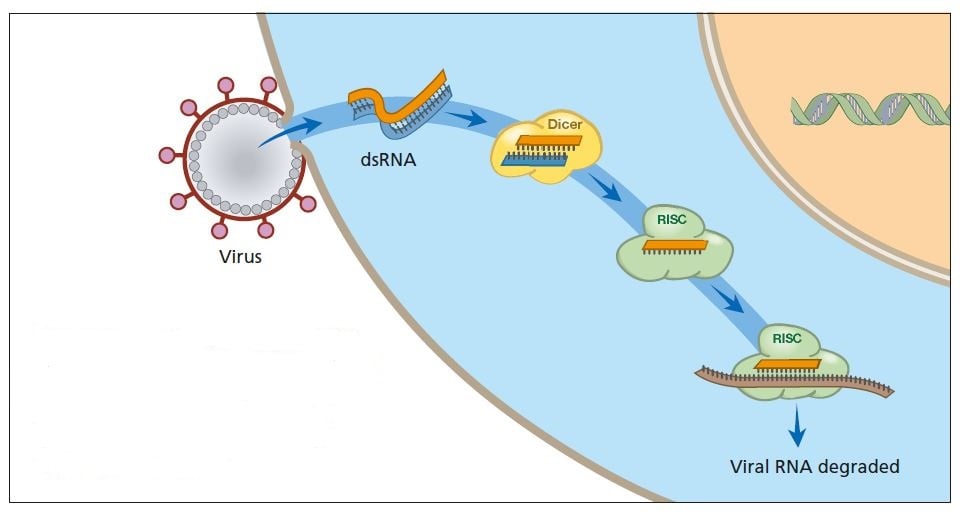
Primitive organisms use RNA interference to protect themselves from virus infections. Double-stranded RNA from a virus activates the protein complexes Dicer and RISC and the infection is stopped.
© The Nobel Assembly at Karolinska Institutet
© The Nobel Assembly at Karolinska Institutet
Protection against jumping genes
Most organisms have moveable segments of DNA, transposons. These DNA sequences are also called jumping genes because on rare occasions they can move around in the genome. Transposons – like mutations – contribute to evolution by increasing genetic variation. But if a transposon happens to end up in an inappropriate place, for instance in an important gene, it can be harmful.
Certain stages of this “jumping” in the genome require that transposon DNA is copied as RNA molecules. The RNA that is formed can be double-stranded and can then be broken down by RNA interference. In this way, RNA interference protects the genome against the damaging effects of transposons.
Molecules tailor-made in the laboratory
In addition to its natural functions, the RNA interference discovered by Fire and Mello opens the way to exciting new possibilities for genetic engineering. RNA interference has already become an important tool for biological and medical research; for instance, it is widely used as a powerful method for mapping gene functions.
In the laboratory, researchers can tailor RNA molecules – silencing RNAs – that activate breakdown of endogenous mRNAs (see figure below). When silencing RNA molecules enter the cell they activate RNA interference. Endogenous mRNA molecules that bind to the added RNA are destroyed. Researchers hope this principle can be applied in many contexts – plant breeding, for instance – and eventually be used to combat serious diseases (see below).
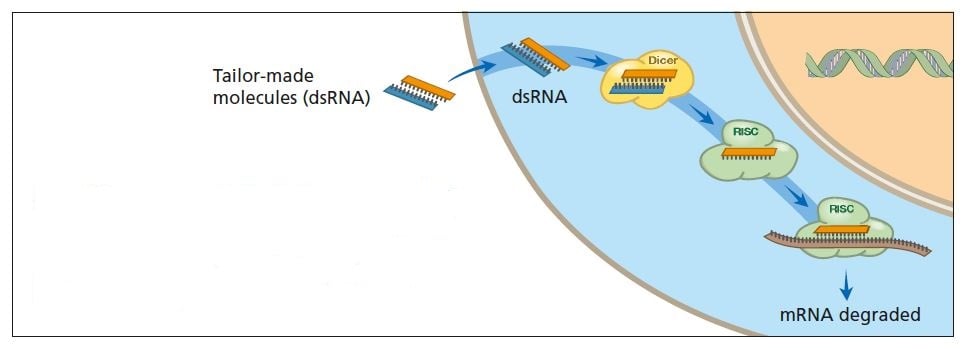
Scientists can now “tailor” double-stranded RNA molecules to activate RNA interference. This makes it possible to turn off specific genes. In the future it may be possible to use this technique to treat diseases.
© The Nobel Assembly at Karolinska Institutet
© The Nobel Assembly at Karolinska Institutet
Medical applications?
The discovery of RNA interference raised hopes for improved treatment of many different disorders, including viral infections, cardiovascular diseases, cancer and metabolic disorders. In many diseases, certain genes are over-expressed (that is, overactive), and it might be possible to alleviate these disorders by suppressing the activity of specific genes.
It is conceptually attractive to use RNA interference to treat diseases: RNA interference is a natural mechanism and double-stranded RNA is an endogenous substance. RNA interference – unlike antisense techniques – has also been shown to give reproducible results. In addition, double-stranded RNA molecules can easily be synthesized.
So far no drugs based on RNA interference have been approved, but successful animal experiments have been performed and several substances are being tested in clinical trials in humans. For example, double-stranded RNA is being tested as a means to treat agerelated degeneration of the fovea (neovascular or “wet” macular degeneration). This condition is common among elderly people and can severely reduce the eyesight. Blood vessels grow into and damage the fovea, the part of the retina that gives the sharpest image. One important cause of vessel growth is a substance called VEGF (vascular endothelial growth factor). If double-stranded RNA corresponding to the mRNA for VEGF is injected into the eye, this can decrease the amount of the growth factor and prevent new vessels from invading the fovea.
Promising experiments
RNA interference is also being tested as a method to combat RS virus, which can cause severe respiratory infections in small children. The principle behind the treatment is that when the child inhales double-stranded RNA, viruses in the lung will be inactivated and the infection will be terminated. To date, one study has been carried out on healthy human volunteers and the treatment did not have any serious side-effects.
Many RNA interference studies on animals and in cell cultures are also currently ongoing. In a recent study on animals (rodents and monkeys), double-stranded RNA was successfully used to silence a gene that leads to high cholesterol levels. Experiments have also been performed, in human cells, where RNA interference has been found to reduce the activity of the AIDS virus HIV.
Although many experiments with RNA interference have yielded promising results, quite a few problems remain to be solved to make this treatment work optimally. For example, we have to be able to get the double-stranded RNA molecules into the right type of cell and the right number of those cells. It must also be possible to control treatment so the double-stranded RNA is active for the appropriate length of time.
Andrew Fire and Craig Mello have been awarded the Nobel Prize for their discovery of a central mechanism of gene regulation. The future will show whether their finding will contribute to the development of new treatment strategies – it looks promising.
The Laureates
Andrew Fire, born in 1959, is a US citizen. Since 2003 he has been professor of Pathology and Genetics at Stanford University School of Medicine, Stanford, California, USA.
In 1983 he took his PhD in Biology at the Massachusetts Institute of Technology, Cambridge, Massachusetts, USA. He began his research on the nematode C. elegans during his time as visiting scientist in Cambridge, England, at the laboratory of Sydney Brenner (Nobel Laureate 2002). When Fire and Mello made their key discoveries about RNA interference, Fire was working at the Carnegie Institution of Washington.
Craig Mello, born in 1960, is a US citizen and a professor of Molecular Medicine. Since 1994 he has worked within the Program in Molecular Medicine, University of Massachusetts Medical School, Worcester, Massachusetts, USA. He is also a Howard Hughes Medical Institute Investigator.
In 1990 he took his PhD in Cellular and Developmental Biology at Harvard University, Boston, Massachusetts. Before he moved to the University of Massachusetts Medical School in Worcester, he worked at the Fred Hutchinson Cancer Research Center.
Previous Nobel Prizes in this research field
The mechanisms underlying how information in the DNA of the cell nucleus is transformed into proteins and how our genes are regulated have been revealed step by step by many scientists. This year’s award touches on aspects of many earlier Nobel Prizes in Physiology or Medicine.
1933: Thomas Morgan, USA, “for his discoveries concerning the role played by the chromosome in heredity”.
1959: Severo Ochoa and Arthur Kornberg, USA, “for their discovery of the mechanisms in the biological synthesis of ribonucleic acid and deoxyribonucleic acid”.
1962: Francis Crick, United Kingdom, James Watson, USA, and Maurice Wilkins, United Kingdom, “for their discoveries concerning the molecular structure of nucleic acids and its significance for information transfer in living material”.
1965: François Jacob, André Lwoff and Jacques Monod, France, “for their discoveries concerning genetic control of enzyme and virus synthesis”.
1968: Robert Holley, Har Gobind Khorana and Marshall Nirenberg, USA, “for their interpretation of the genetic code and its function in protein synthesis”.
1978: Werner Arber, Switzerland, Daniel Nathans and Hamilton Smith, USA, “for the discovery of restriction enzymes and their application to problems of molecular genetics”.
1983: Barbara McClintock, USA, “for her discovery of mobile genetic elements”.
1993: Richard Roberts, United Kingdom, and Phillip Sharp, USA, “for their discoveries of split genes”.
In addition to these Nobel Prizes in Physiology or Medicine, research in this field has been awarded two Nobel Prizes in Chemistry.
1989: Sidney Altman and Thomas Cech, USA, “for their discovery of catalytic properties of RNA”.
2006: Roger Kornberg, USA, “for his studies of the molecular basis of eukaryotic transcription”.
For further reading
A. Fire, S.Q. Xu, M.K. Montgomery, S.A. Kostas, S.E. Driver & C.C. Mello: Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998; 391:806-811.
The editorial committee for this year’s popular presentation of the Nobel Prize in Physiology or Medicine included the following, who are professors at Karolinska Institutet and members of the Nobel Assembly:
Bertil Daneholt, Göran K. Hansson, Hans Jörnvall and Nils-Göran Larsson
Text: Anders Nystrand, MD and medical writer
Illustrations: Annika Röhl, graphic editor
Translation: Janet Holmén, editor
© 2006 The Nobel Committee for Physiology or Medicine, Karolinska Institutet, SE-171 77 Stockholm, Sweden.
Nobel Prizes and laureates
Six prizes were awarded for achievements that have conferred the greatest benefit to humankind. The 14 laureates' work and discoveries range from quantum tunnelling to promoting democratic rights.
See them all presented here.
