Advanced information
Scientific Background:
Discoveries of Molecular Mechanisms Controlling the Circadian Rhythm (pdf)
![]()
Scientific Background
Discoveries of Molecular Mechanisms Controlling the Circadian Rhythm
The 2017 Nobel Prize in Physiology or Medicine is awarded to Jeffrey C. Hall, Michael Rosbash and Michael W. Young for their discoveries of molecular mechanisms that control circadian rhythms. Circadian rhythms are driven by an internal biological clock that anticipates day/night cycles to optimize the physiology and behavior of organisms. Observations that organisms adapt their physiology and behavior to the time of the day in a circadian fashion have been documented for a long time, but the existence of an endogenous circadian clock would only finally become established well into the 20th century. In 1971, Seymour Benzer and Ronald Konopka identified mutants of the fruit fly Drosophila that displayed alterations in the normal 24h cycle of pupal eclosion and locomotor activity. Experiments suggested that the mutations involved the same gene, later named period. A decade later, Hall and Rosbash, collaborating at Brandeis University, and Young, at Rockefeller University, isolated and molecularly characterized the period gene. However, its structure and sequence did not immediately suggest a molecular mechanism for the circadian clock. A series of breakthroughs, including the identification of other genes that partner with period, from Hall, Rosbash and Young eventually led to the notion of a Transcription- Translation Feedback Loop (TTFL). In this mechanism, the transcription of period and its partner gene timeless are repressed by their own gene products – the PERIOD (PER) and TIME- LESS (TIM) proteins, generating an autonomous oscillation. At the time, a transcriptional mechanism was not obvious, and the discovery of the self- sustained circadian TTFL was a new paradigm. Further studies revealed a series of interlocked transcription-translation feedback loops, together with a complex network of reactions. These involve regulated protein phosphorylation and degradation of TTFL components, protein complex assembly, nuclear translocation and other post-translational modifications, generating oscillations with a period of ~24 hours. Circadian oscillators within individual cells respond differently to entraining signals and control various physiological outputs, such as sleep patterns, body temperature, hormone release, blood pressure, and metabolism. The seminal discoveries by Hall, Rosbash and Young have revealed a crucial physiological mechanism explaining circadian adaptation, with important implications for human health and disease.
What makes us tick?
A key feature of life on Earth is its capacity to adapt to the environment. Different geographical locations have different environments and organisms adapt to the conditions that are prevalent at their location to enhance their survival. However, at any given location, profound changes in environmental light and temperature occur daily as a consequence of the rotation of the Earth on its axis. To adapt to such changes, most organisms have evolved an internal biological clock that anticipates day/night cycles and helps them optimize their physiology and behavior. This internally generated daily rhythm is known as “circadian”, from the Latin words circa meaning “around” and dies meaning “day”. Circadian rhythms are ancient and conserved throughout evolution. They are known to exist in life forms from unicellular cyanobacteria and protozoans to all multicellular organisms, including fungi, plants, insects, rodents and humans. The building blocks of a circadian system consist of a self-sustained 24-hour rhythm generator or oscillator, setting or entraining mechanisms that link the internal oscillator to external stimuli (referred to as zeitgebers, i.e. timekeepers), such as light, and output mechanisms to allow the timely scheduling of physiological processes.
From rhythms to clocks
Observations that organisms adapt their physiology and behavior to the time of the day in a circadian fashion have been documented for a long time and are commonly agreed to have begun with the observation of leaf and flower movements in plants. For example, the leaves of mimosa plants close at night and open during the day. In 1729, the French astronomer Jean Jacques d’Ortous de Mairan placed a mimosa plant in the dark and observed that the leaves still opened and closed rhythmically at the appropriate time of the day, suggesting an endogenous origin of the daily rhythm (Figure 1). About two hundred years later, the German plant physiologist and pioneer of circadian rhythm research, Erwin Bünning, painstakingly connected the leaves of a bean plant to a kymograph and recorded the movements of the leaves during normal day/night cycles and under constant light conditions. He observed that the rhythm of leaf movement persisted. The question of whether circadian behaviors in plants and animals were governed by an endogenous clock, or were a mere reaction to external stimuli of a circadian nature, would be hotly debated for decades. Eventually, the existence of an endogenous circadian clock would finally become established well into the 20th century.
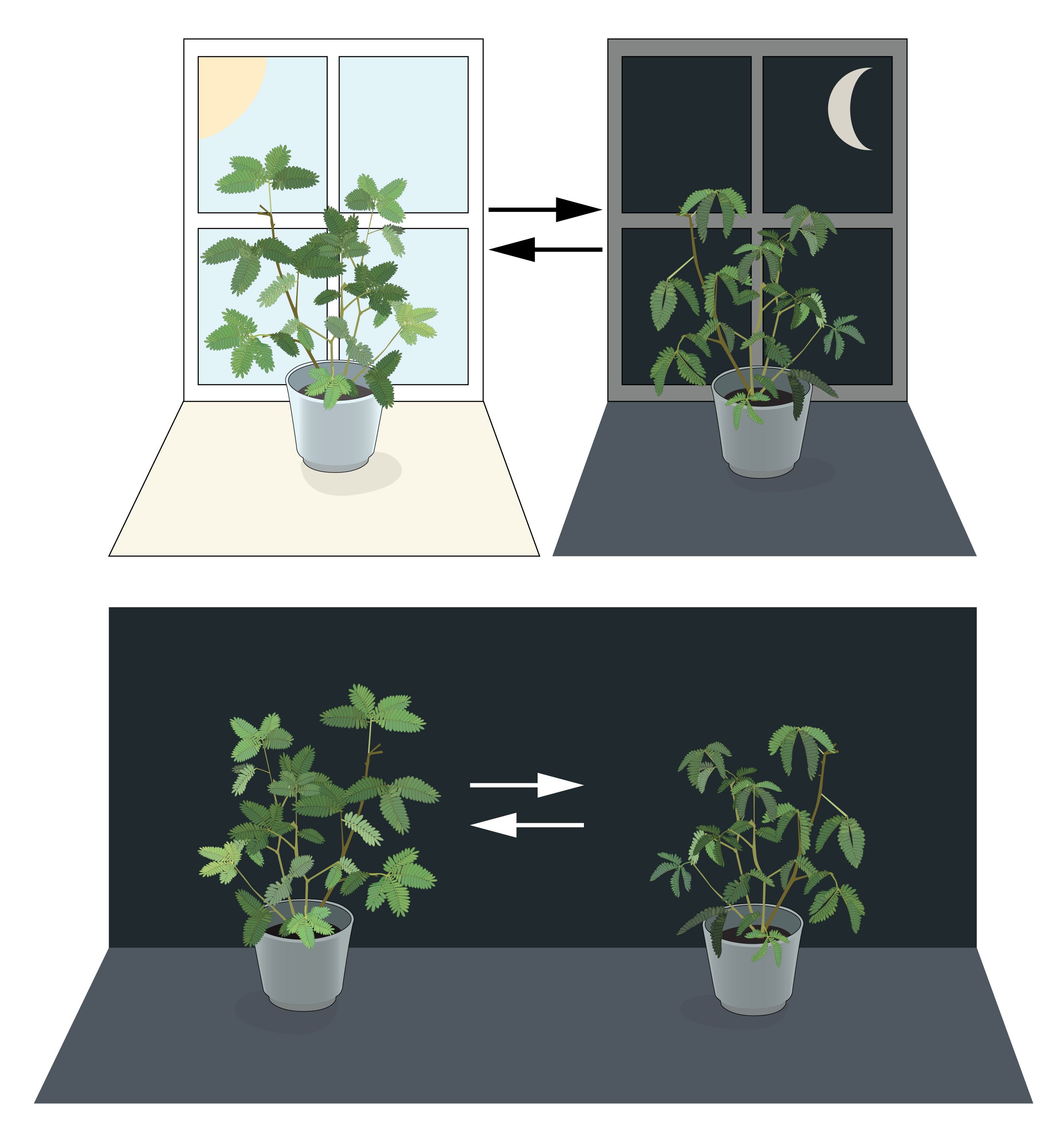
Figure 1. An internal biological clock.
Leaves of mimosa plants open towards the sun during daytime and close at dusk. Jean Jacques d’Ortous de Mairan placed a mimosa plant in constant dark and found that the leaves continued to follow their daily rhythm for several days. This suggested that mimosa plants have a cell autonomous clock that can maintain the biological rhythm even under constant conditions.
Heritability of circadian rhythms and clock genes
With time, many relevant physiological properties besides periodic leaf movements were found to be controlled by the physiological clock and the inheritance of circadian rhythms began to be considered as the product of natural selection. Erwin Bünning’s classical studies in the 1930s showed that circadian rhythms in plants can be inherited despite parent plants being exposed to non-circadian light periods and that crosses between strains with varying periods yielded plants with intermediate periods. By the mid- 1960s, a community of chronobiology researchers investigating biological clocks was well established and the concept of clock genes began to be contemplated.
It was at about this time that Seymour Benzer and his student Ronald Konopka, working at the California Institute of Technology, embarked on studies to identify mutant fruit flies with altered circadian phenotypes. Unlike several geneticists and behavioral scientists of the time, Benzer firmly believed that specific behaviors may be influenced by the action of single genes and that it would be possible to demonstrate this by isolating organisms with altered behavior carrying mutations in individual genes. Using a classical chemical-based mutagenesis strategy, Benzer and Konopka isolated three different strains of mutant flies showing alterations in the normal 24h cycle of pupal eclosion and locomotor activity (Konopka and Benzer, 1971). One mutant was arrhythmic, another had a shorter period of 19h, and a third had a longer period of 28h. Mapping experiments, using the genetic markers known at the time, roughly localized all three mutants to the same region of the X chromosome of the fruit fly. Importantly, complementation tests suggested that the three mutations involved the same gene, later named period. Based on this, Benzer and Konopka presciently predicted that the arrhythmic mutant would carry a nonsense mutation that inactivated the gene, and that the mutants with longer and shorter periods would carry missense mutations that somehow altered the function of the gene product in opposite ways. Later work showed both predictions to be correct. Although Benzer would move on to other topics, Konopka continued working on the period locus, mapping its chromosomal position with greater precision. However, the period gene would not be molecularly cloned and sequenced until the mid- 1980s through the work of Jeffrey Hall and Michael Rosbash, collaborating at Brandeis University, and Michael Young, at Rockefeller University (Bargiello and Young, 1984; Bargiello et al., 1984; Reddy et al., 1984; Zehring et al., 1984). The first clock gene was thereby isolated and its structure was molecularly characterized. However, neither the original genetic identification of period nor the cloning and sequencing of its cDNA pointed to a molecular mechanism for the circadian clock.
The Transcription-Translation Feedback Loop
In the years following the cloning of period, several models were proposed to explain how its protein product PER might function to produce circadian oscillations. A “membrane gradient” model was proposed in which PER was envisioned to function like a pump to build a gradient across the membrane which, upon reaching a threshold, gets dissipated through light-sensitive channels. In another model, the PER protein was proposed to be a proteoglycan that brings cells together, thereby facilitating the formation of inter-cellular connections through gap junctions. A series of breakthroughs were finally made possible with the availability of reliable PER antibodies. First was the discovery from the Hall and Rosbash laboratories of a 24h cycle in the abundance of PER protein in neurons of the fly brain, with a peak during the night (Siwicki et al., 1988). The mRNA encoded by the period gene also showed circadian cycles of abundance in fly brain, showing that the cycling of PER protein resulted from the cycling of period mRNA. Intriguingly, the peak of period mRNA levels occurred early in the night, several hours before the peak in PER protein abundance (Hardin et al., 1990). Importantly, a nonsense mutant of period was incapable of generating oscillating levels of period mRNA, but wild type PER protein could rescue cyclic mRNA expression. Based on these observations, the negative autoregulatory feedback model was born, whereby the accumulation of PER protein resulted in the attenuation of period mRNA expression (Hardin et al., 1990). Subsequently, PER protein was found to be a nuclear protein and to shuttle between the cell nucleus and the cytoplasm in a temporally regulated manner, providing support for the idea that PER protein was a transcriptional regulator of some kind (Liu et al., 1992). By a new forward screen, Young discovered timeless, an additional gene influencing the circadian clock (Myers et al., 1995; Sehgal et al., 1995). In a string of subsequent discoveries the Young laboratory found that the levels of timeless mRNA also cycled with a 24h period, and that TIM could bind directly to PER, affecting its nuclear localization and abundance by blocking PER degradation (Gekakis et al., 1995; Sehgal et al., 1994; Vosshall et al., 1994). Importantly, the cycle of period expression was abolished in timeless mutant flies and, conversely, circadian cycles in timeless expression were lost in period mutant flies (Sehgal et al., 1994; 1995). These advances consolidated the basic conceptual framework of the TTFL as a mechanism for promoting circadian cycles in the autoregulation of clock genes (Figure 2A). At the time, a transcriptional mechanism was not obvious and, as noted above, different alternatives were being considered. Thus, the discovery of the self-sustained circadian TTFL represents a new paradigm.
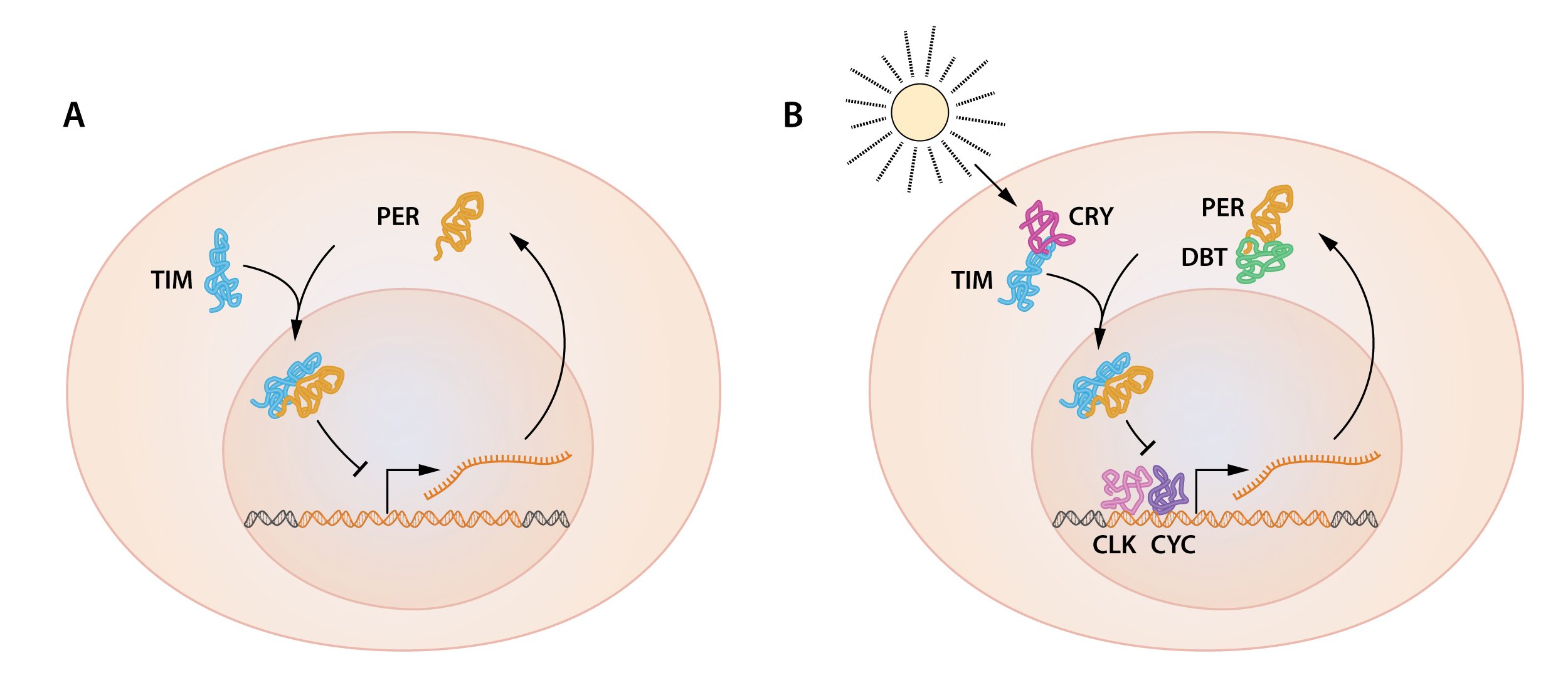
Figure 2. A simplified illustration of the feedback regulation of the period gene.
A) Both period mRNA and PER protein oscillate, with PER protein accumulating several hours after the peak in period mRNA. PER protein localizes in the nucleus, and the period gene activity oscillates as a result of PER protein feedback inhibition of its own gene. B) Additional proteins are essential for the oscillation of the period gene. TIM protein, encoded by the timeless gene is also oscillating and interacts with PER protein. The interaction is critical for PER protein nuclear accumulation and repression of the period gene. DBT protein is encoded by the doubletime gene. DBT is a protein kinase that phosphorylates PER, leading to PER protein degradation. DBT-mediated PER protein degradation contributes to the delay between period mRNA and PER protein accumulation. CLK and CYK, encoded by the clock and cycle genes, are two transcription factors that activate the period gene.
The mechanism by which period and timeless transcription was activated remained unknown. This question was resolved with the discovery of the clock and cycle genes (Allada et al., 1998; Rutila et al., 1998). The Clock gene was first identified in the mouse, by Joseph Takahashi (King et al., 1997). The gene products, CLOCK (CLK) and CYCLE (CYC) interact with each other, contain basic helix-loop-helix (bHLH) motifs, and bind to specific elements in the period and timeless genes, thereby positively regulating their transcription. Later studies would show that TIM and PER act as negative regulators of CLK activity, and by this, the circadian feedback loop is closed (Darlington et al., 1998).
Current working models of the circadian molecular clockwork are highly complex and include many additional components which, collectively, contribute to its robustness and circadian periodicity (Hardin, 2011). Importantly, as transcription and translation reactions are typically rapid, substantial delays must be imposed on the core TTFL mechanism to generate 24h oscillations. This is achieved by a complex network of reactions involving regulated protein phosphorylation and degradation of TTFL components, protein complex assembly, nuclear translocation and other post-translational modifications (Hardin, 2011). A key observation demonstrating the underlying mechanism for such a delay came from the discovery by Young of the doubletime gene, encoding a kinase DOUBLETIME (DBT) that phosphorylates PER and increases its degradation (Price et al., 1998). Additional proteins integrate environmental inputs that can entrain the clock (Figure 2B). For instance, light can activate the protein product of the cryptochrome cry gene (CRY) and promote its binding to TIM, leading to its degradation in the proteasome (Ceriani et al., 1999; Emery et al., 1998). When morning arrives, TIM is degraded, leaving PER vulnerable to phosphorylation by DBT and subsequent degradation.
Circadian clocks in other organisms
TTFL mechanisms are also an underlying principle of circadian clocks in other multicellular organisms, including humans. Several homologues of the core clock proteins in Drosophila, including CLK and PER, play similar roles in mammalian circadian timekeeping (Papazyan et al., 2016). Although plants mainly use transcription factors that are not homologous to those in the Drosophila circadian clock, TTFLs is the unifying main principle (Nohales and Kay, 2016). However, in cyanobacteria, a different type of transcription-independent circadian oscillator has been described that depends on sequential protein phosphorylation events (Tomita et al., 2005). Remarkably, a circadian rhythm can be reconstituted in vitro using purified cyanobacteria clock proteins and ATP (Nakajima et al. 2005). A transcription-independent oscillation resulting from hyperoxidation of peroxiredoxin has also been described in eukaryotes, including in human erythrocytes (O’Neill et al., 2011; Ray and Reddy, 2016). The physiological relevance of such TTFL- independent oscillations is unknown. Nevertheless, these results suggest that additional mechanisms for generating circadian oscillations may also exist in mammalian cells.
Entrainment and synchronization of biological clocks
The circadian program is regulated at both a central and peripheral level. In mammals, the central pacemaker is located in the suprachiasmatic nucleus (SCN) of the hypothalamus and functions as the master circadian clock. The retina receives photic input and relays this information to the SCN, which synchronizes its own neuronal cellular clocks. The central clock regulates circadian rhythms across the entire body via humoral factors and the peripheral autonomic nervous system. However, the capacity for circadian gene expression is widespread throughout the body and most peripheral organs and tissues can express circadian oscillations in isolation (Balsalobre et al., 1998). Thus, the circadian system of an animal resembles a clock shop rather than a single clock. This has raised the question of how so many clocks can be effectively synchronized (Mohawk et al., 2012).
Peripheral clocks can be synchronized both by the SCN and by environmental cues, including feeding, physical activity and temperature. Peripheral clocks in different tissues control relevant physiological outputs, such as glucose production, fat storage and release of hormones (Panda, 2016). These, in turn, function as time- keeping cues for clocks in tissues throughout the body, ultimately feeding back to the SCN. Thus, the circadian system of an organism is a web of interconnected oscillators and feedback loops. The relationship between the central and peripheral clocks, and the multiple ways by which local and external cues affect them, is an active area of research open to new discoveries.
Circadian biology and human health
Chronobiology has an impact on many aspects of our physiology. For example, circadian clocks help to regulate sleep patterns, feeding behavior, hormone release, blood pressure and body temperature (Figure 3). Molecular clocks also play critical roles locally in many tissues. Ablation of clock genes in animal models results in arrhythmic production of hormones, such as corticosterone and insulin (Son et al., 2008). Clock genes also exert a profound influence on metabolism through the control of gluconeogenesis, insulin sensitivity and systemic oscillation of blood glucose (Panda, 2016). Sleep is vital for normal brain function and circadian dysfunction has been linked to sleep disorders, as well as depression, bipolar disorder, cognitive function, memory formation and some neurological diseases (Gerstner and Yin, 2010). In rare cases, sleep phase disorders are due to mutations in circadian clock genes resulting in advanced or delayed sleep-wake cycles (Patke et al., 2017; Toh et al., 2001). Studies have indicated that chronic misalignment between our lifestyle and the rhythm dictated by our endogenous circadian clock may be associated with increased risk for various diseases including cancer, neurodegenerative diseases, metabolic disorders and inflammation. Efforts are underway to develop approaches in chronobiology and pharmacology to modify the period, phase or amplitude of circadian clocks to improve human health (Hirota and Kay, 2015).
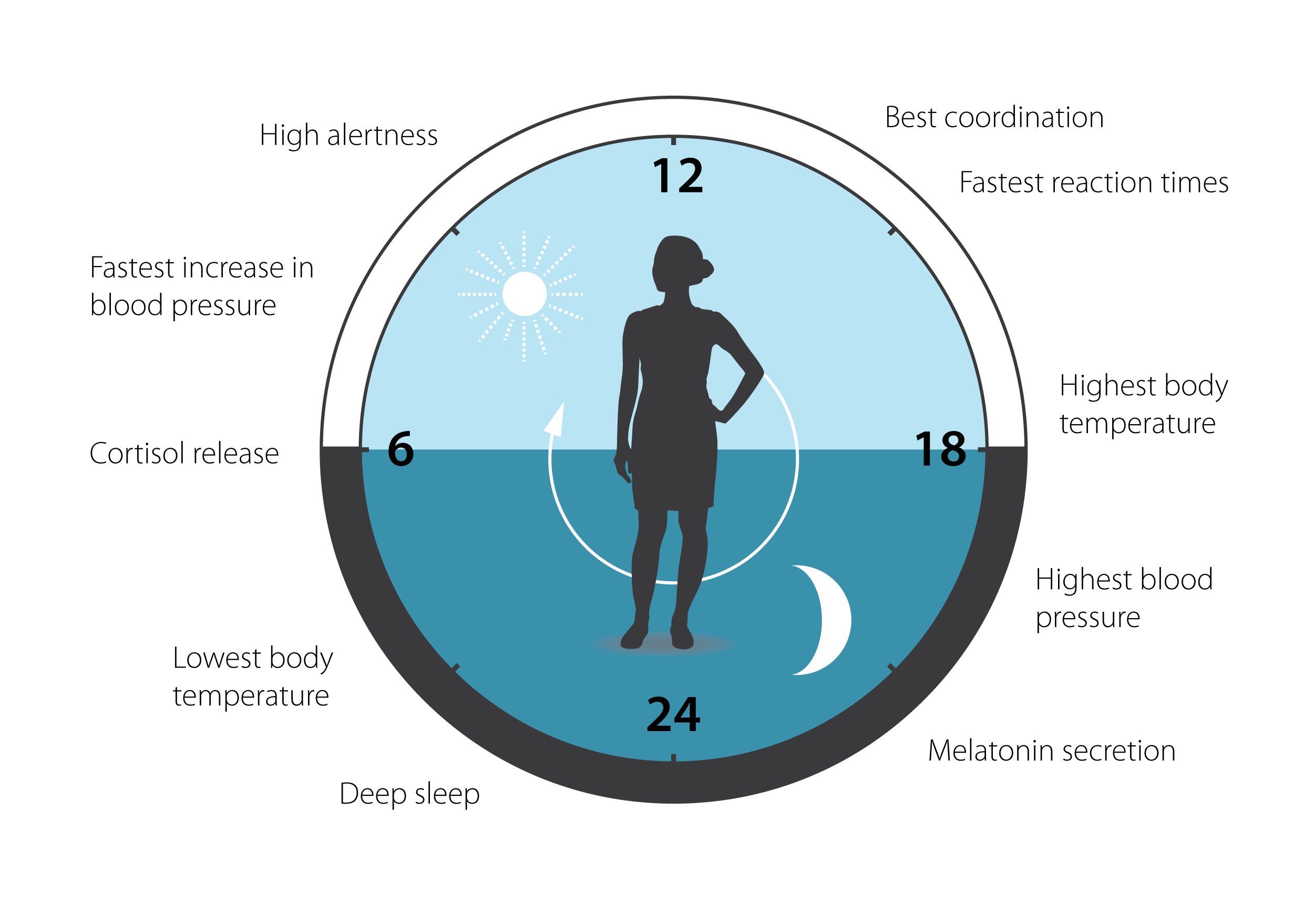
Figure 3. The circadian clock has an impact on many aspects of our physiology.
This clock helps to regulate sleep patterns, feeding behavior, hormone release, blood pressure and body temperature. A large proportion of our genes are regulated by the clock.
Conclusions
The discovery of self-sustained transcription/ translation feedback loops as the central component of the molecular mechanism by which clock genes control circadian oscillations in cells and tissues has led to a new paradigm in our understanding of how organisms anticipate and adapt to the regular daily environmental cues such as light. Since the seminal discoveries by the three laureates, elucidating a fundamental physiological mechanism, circadian biology has developed into a vast and highly dynamic research field, with important implications for our health and wellbeing.
Carlos Ibáñez, PhD
Professor of Neuroscience, Karolinska Institutet
Adjunct Member of the Nobel Committee
Member of the Nobel Assembly
Illustration: Mattias Karlén
References
Allada, R., White, N.E., So, W.V., Hall, J.C., and Rosbash, M. (1998). A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell 93, 791– 804.
Balsalobre, A., Damiola, F., and Schibler, U. (1998). A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937.
Bargiello, T.A., and Young, M.W. (1984). Molecular genetics of a biological clock in Drosophila. Proc Natl Acad Sci USA 81, 2142–2146.
Bargiello, T.A., Jackson, F.R., and Young, M.W. (1984). Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature 312, 752–754.
Ceriani, M.F., Darlington, T.K., Staknis, D., Más, P., Petti, A.A., Weitz, C.J., and Kay, S.A. (1999). Light- dependent sequestration of TIMELESS by CRYPTOCHROME. Science 285, 553–556.
Darlington, T.K., Wager-Smith, K., Ceriani, M.F., Staknis, D., Gekakis, N., Steeves, T.D., Weitz, C.J., Takahashi, J.S., and Kay, S.A. (1998). Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 280, 1599–1603.
Emery, P., So, W.V., Kaneko, M., Hall, J.C., and Rosbash, M. (1998). CRY, a Drosophila clock and light- regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95, 669–679.
Gekakis, N., Saez, L., Delahaye-Brown, A.M., Myers, M.P., Sehgal, A., Young, M.W., and Weitz, C.J. (1995). Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL. Science 270, 811–815.
Gerstner, J.R., and Yin, J.C.P. (2010). Circadian rhythms and memory formation. Nat Rev Neurosci 11, 577–588.
Hardin, P.E., Hall, J.C., and Rosbash, M. (1990). Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343, 536–540.
Hardin, P.E. (2011). Molecular genetic analysis of circadian timekeeping in Drosophila. Adv. Genet. 74, 141–173.
Hirota, T., and Kay, S.A. (2015). Identification of small-molecule modulators of the circadian clock. Meth. Enzymol. 551, 267–282.
Ishiura, M., Kutsuna, S., Aoki, S., Iwasaki, H., Andersson, C.R., Tanabe, A., Golden, S.S., Johnson, C.H., and Kondo, T. (1998). Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281, 1519–1523.
King, D.P., Zhao, Y., Sangoram, A.M., Wilsbacher, L.D., Tanaka, M., Antoch, M.P., Steeves, T.D., Vitaterna, M.H., Kornhauser, J.M., Lowrey, P.L., et al. (1997). Positional cloning of the mouse circadian clock gene. Cell 89, 641–653.
Konopka, R.J., and Benzer, S. (1971). Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA 68, 2112–2116.
Liu, X., Zwiebel, L.J., Hinton, D., Benzer, S., Hall, J.C., and Rosbash, M. (1992). The period gene encodes a predominantly nuclear protein in adult Drosophila. J Neurosci 12, 2735–2744.
Mohawk, J.A., Green, C.B., and Takahashi, J.S. (2012). Central and peripheral circadian clocks in mammals. Annu Rev Neurosci 35, 445–462.
Myers, M.P., Wager-Smith, K., Wesley, C.S., Young, M.W., and Sehgal, A. (1995). Positional cloning and sequence analysis of the Drosophila clock gene, timeless. Science 270, 805–808.
Nakajima, M., Imai, K., Ito, H., Nishiwaki, T., Murayama, Y., Iwasaki, H., Oyama, T., and Kondo, T. (2005). Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science, 308, 414-415
Nohales, M.A., and Kay, S.A. (2016). Molecular mechanisms at the core of the plant circadian oscillator. Nat Struct Mol Biol 23, 1061–1069.
O’Neill, J.S., van Ooijen, G., Dixon, L.E., Troein, C., Corellou, F., Bouget, F.-Y., Reddy, A.B., and Millar,
A.J. (2011). Circadian rhythms persist without transcription in a eukaryote. Nature 469, 554–558. Panda, S. (2016). Circadian physiology of metabolism. Science 354, 1008–1015.
Papazyan, R., Zhang, Y., and Lazar, M.A. (2016). Genetic and epigenomic mechanisms of mammalian circadian transcription. Nat Struct Mol Biol 23, 1045–1052.
Patke, A., Murphy, P.J., Onat, O.E., Krieger, A.C., Özçelik, T., Campbell, S.S., and Young, M.W. (2017). Mutation of the Human Circadian Clock Gene CRY1 in Familial Delayed Sleep Phase Disorder. Cell 169, 203–215.e213.
Price, J.L., Blau, J., Rothenfluh, A., Abodeely, M., Kloss, B., and Young, M.W. (1998). double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94, 83–95.
Ray, S., and Reddy, A.B. (2016). Cross-talk between circadian clocks, sleep-wake cycles, and metabolic networks: Dispelling the darkness. Bioessays 38, 394–405.
Reddy, P., Zehring, W.A., Wheeler, D.A., Pirrotta, V., Hadfield, C., Hall, J.C., and Rosbash, M. (1984). Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell 38, 701–710.
Rutila, J.E., Suri, V., Le, M., So, W.V., Rosbash, M., and Hall, J.C. (1998). CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93, 805–814.
Sehgal, A., Price, J.L., Man, B., and Young, M.W. (1994). Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science 263, 1603–1606.
Sehgal, A., Rothenfluh-Hilfiker, A., Hunter-Ensor, M., Chen, Y., Myers, M.P., and Young, M.W. (1995). Rhythmic expression of timeless: a basis for promoting circadian cycles in period gene autoregulation. Science 270, 808–810.
Siwicki, K.K., Eastman, C., Petersen, G., Rosbash, M., and Hall, J.C. (1988). Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron 1, 141–150.
Son, G.H., Chung, S., Choe, H.K., Kim, H.-D., Baik, S.-M., Lee, H., Lee, H.-W., Choi, S., Sun, W., Kim,
H., et al. (2008). Adrenal peripheral clock controls the autonomous circadian rhythm of glucocorticoid by causing rhythmic steroid production. Proceedings of the National Academy of Sciences 105, 20970– 20975.
Toh, K.L., Jones, C.R., He, Y., Eide, E.J., Hinz, W.A., Virshup, D.M., Ptácek, L.J., and Fu, Y.H. (2001). An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291, 1040– 1043.
Tomita, J., Nakajima, M., Kondo, T., and Iwasaki, H. (2005). No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307, 251-254.
Vosshall, L.B., Price, J.L., Sehgal, A., Saez, L., and Young, M.W. (1994). Block in nuclear localization of period protein by a second clock mutation, timeless. Science 263, 1606–1609.
Zehring, W.A., Wheeler, D.A., Reddy, P., Konopka, R.J., Kyriacou, C.P., Rosbash, M., and Hall, J.C. (1984). P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster. Cell 39, 369–376.
Nobel Prizes and laureates
Six prizes were awarded for achievements that have conferred the greatest benefit to humankind. The 12 laureates' work and discoveries range from proteins' structures and machine learning to fighting for a world free of nuclear weapons.
See them all presented here.
