Press release

NOBELFÖRSAMLINGEN KAROLINSKA INSTITUTET
THE NOBEL ASSEMBLY AT THE KAROLINSKA INSTITUTE
11 October 1993
The Nobel Assembly at the Karolinska Institute has today decided to award the 1993 Nobel Prize in Physiology or Medicine jointly to
Richard J. Roberts and Phillip A. Sharp
for their discovery of “split genes”.
Summary
Our knowledge regarding the genetic material, the genes, has increased dramatically during the last forty years due to achievements in the area of molecular biology. During the first decades, studies on simple organisms, in particular bacteria and bacterial viruses, dominated. A gene was conceived as a continuous segment within the very long double-stranded DNA molecules, the chemical substance of heredity. This simple picture of gene structure completely changed when Richard J. Roberts and Phillip A. Sharp in 1977 independently discovered that genes could be discontinuous, that is, a given gene could be present in the genetic material (DNA) as several, well-separated segments. As their experimental model system, both Roberts and Sharp used a common cold-causing virus, called adenovirus, whose genes display important similarities to those in higher organisms. Shortly thereafter it could be shown by several researchers that split genes are frequent in higher organisms, including man.
Roberts’ and Sharp’s discovery has changed our view on how genes in higher organisms develop during evolution. The discovery also led to the prediction of a new genetic process, namely that of splicing, which is essential for expressing the genetic information. The discovery of split genes has been of fundamental importance for today’s basic research in biology, as well as for more medically oriented research concerning the development of cancer and other diseases.
The genetic material
During the last forty years our knowledge of how the genetic material, the genes, governs the basic activities of life has increased dramatically. This is due to progress made within molecular biology, the area in science which explores biological phenomena and structures at the molecular level. Many of the most important discoveries within this area have been awarded a Nobel Prize. Examples include the discovery of how the nucleic acid DNA, the chemical substance of heredity, is built (1962), how the synthesis of nucleic acids takes place (1959), how the activity of genes is regulated (1965) and what the genetic code looks like (1968). This knowledge evolved primarily through studies of simple organisms such as bacteria and viruses infecting bacteria.
The general concept prevailing during the mid 1970s regarding the hereditary material and its function can be summarized as follows. A gene exists as a continuous stretch (segment) within a long, double-stranded DNA molecule. When the gene is activated, its information is copied into a single-stranded RNA molecule, called messenger RNA, which translates the information into a protein (figure 1A).
This simple picture of the sequence of events radically changed through the discovery made in 1977 by Richard J. Roberts, working at the Cold Spring Harbor Laboratory on Long Island, New York, and Phillip A. Sharp, working at the Massachusetts Institute of Technology in Cambridge, USA. They found that an individual gene can comprise not only one but several DNA segments separated by irrelevant DNA (figure 1B). Such discontinuous genes exist in organisms more complex than those studied earlier.
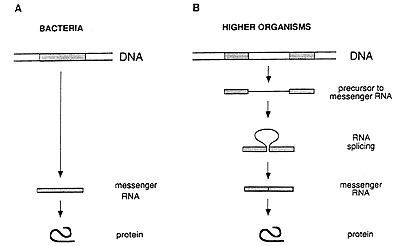 |
Figure 1: Gene structure and the flow of genetic information in bacteria (A) and higher organisms (B). In bacteria, the genetic information is stored as a continuous segment of DNA, and the messenger RNA can immediately direct the synthesis of the corresponding protein. In higher organisms, the gene is usually split, and the messenger RNA has to be processed by splicing before it can be translated into a protein.
How the discovery was made
Roberts and Sharp were studying the genetic material in adenovirus, a virus causing common cold. This virus infects the cells of higher organisms, and its genome has many properties resembling those of the host cell. At the same time, adenovirus has a simple structure, making it a very valuable experimental model for studying genes and their function in higher organisms. The genome of adenovirus consists of one single long DNA molecule. Roberts’ and Sharp’s aim was to determine where in the genome different genes were located.
In biochemical experiments it was shown that one end of an adenovirus messenger RNA did not behave as expected. One of several possible explanations was that the DNA segment corresponding to this end was not located in the immediate vicinity of the rest of the gene. To determine where this segment was located on the long DNA molecule, they used electron microscopy. They surprisingly found that a single RNA molecule corresponded to no less than four well-separated segments in the DNA molecule (figure 2). Roberts and Sharp came to the conclusion that the genetic information in the gene was discontinuously organized in the genome, a conclusion that contradicted the commonly held view regarding the structure of genes. The discovery immediately led to intensive research to find out whether this gene structure is present also in other viruses and in ordinary cells. Very soon after the initial discovery, several researchers could show that a discontinuous (or split) gene structure was common – and in fact the most common gene structure in higher organisms.
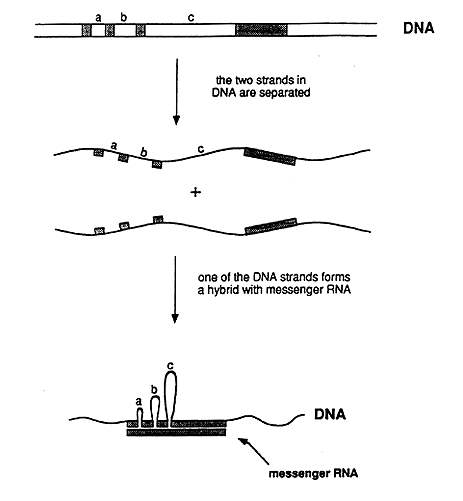
Figure 2: Schematic representation of the experiment that demonstrated that adenovirus DNA contains split genes. The genetic information in the messenger RNA resides in the DNA as four segments, which are separated by three intervening regions (a, b, and c). In the experimentally produced hybrid between one of the DNA strands and the RNA, the intervening sequences in the DNA strand appear as loops, i.e. the corresponding segments lack counterparts in the RNA. The hybrid could be directly visualized in the electron microscope.
The importance of the discovery
A gene may thus consist of several segments, usually termed exons separated by intervening DNA, termed introns. This knowledge has radically changed our view on how the genetic material has developed during the course of evolution. It has long been considered likely that evolution takes place as the result of the accumulation of minor alterations in the genetic material (mutations) resulting in a gradual change.
As a consequence of the discovery that genes are often split, it seems likely that higher organisms in addition to undergoing mutations may utilize another mechanism to speed up evolution: rearrangement (or shuffling) of gene segments to new functional units. This can take place in the germ cells through crossing-over during pairing of chromosomes. This hypothesis seems even more attractive following the discovery that individual exons in several cases correspond to building modules in proteins, so-called domains, to which specific functions can be attributed. An exon in the genome would thus correspond to a particular subfunction in the protein and the rearrangement of exons could result in a new combination of subfunctions in a protein. This kind of process could drive evolution considerably by rearranging modules with specific functions.
The discovery that genes can consist of two or more segments immediately led to a prediction with both surprising and important consequences. The first RNA product synthesized containing both exons and introns has to be “edited” such that the introns are cut out and the remaining exons are joined together to form a shortened RNA molecule. It has now been established that this process indeed takes place, and we have already accumulated detailed information on its nature. The process is called splicing and in higher organisms it represents an additional step in the transfer of information as compared to what usually occurs in lower organisms (figure 1B). The importance of splicing became particularly apparent, when it was found that it is not always the same segments that are identified as exons and are included in the final RNA molecule. In different tissues or developmental stages, the final RNA molecule may be different due to the utilization of alternative exon combinations. Thus, the same DNA region can in many cases determine the structure of many different proteins. The process is called alternative splicing and represents a fundamentally new principle: the genetic message, which gives rise to a particular product, is not definitely established at the stage when the RNA is first synthesized. Instead, it is the splicing pattern that determines the nature of the final product.
Medical aspects
Hereditary diseases are common – their estimated number is today no less than about 5000. Some of them are due to errors in the splicing process. The most studied of such diseases is beta-thalassemia, an anemia prevalent primarily in some Mediterranean countries.
The disease is due to a faulty protein, which forms part of hemoglobin in red blood cells. The protein is called beta-globin. If no or badly functioning beta-globin is made, the life-span of the red blood cells is shortened resulting in anemia. In different patients, small defects in the genetic material have been found, resulting in errors in the splicing process and thus in the synthesis of poorly functioning beta-globin. In the upper part of figure 3 the normal splicing of beta-globin RNA is shown (A). If the globin gene is damaged (marked by an arrow) it may, for example, lead to the formation of a larger than normal exon during splicing (B), or to the formation of a completely new exon (C).
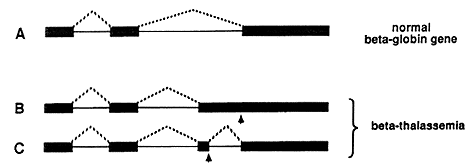 |
Figure 3: Defective splicing causing beta-thalassemia. A normal beta-globin gene is presented in A, and two mutated genes that result in beta-thalassemia are shown in B and C. Arrows mark the position of point mutations. The interrupted lines denote the segments that are being joined during the splicing process. In the healthy individual, three segments are spliced as shown in A. In one of the thalassemia cases, an unusually long third segment is formed (B), while in the second one, an extra segment is produced (C).
Another example showing the connection between disease and the organisation of the genetic material into exons and introns is chronic myeloic leukemia, a type of cancer of the blood. Characteristic for this disease is the presence in tumor cells of a special, shortened chromosome, called the Philadelphia chromosome, named after the city in which it was discovered. This chromosome has arisen in a white blood cell by fusion of one end of chromosome 22 to one end of chromosome 9. At the break-point, a large portion of a cancer gene has been joined to another gene. Here we are thus dealing with two genes, which are now copied into one single RNA molecule. During the splicing process exons from the two genes are spliced to form an RNA molecule that specifies the synthesis of a new protein, a so-called fusion protein. This new protein gives rise to leukemia.
| References |
| B. Alberts et al: Molecular Biology of the Cell. Garland, New York, 1989 |
| P. Chambon: Split Genes. Scientific American 244, 60-71 (1981) |
| J.E. Darnell: RNA. Scientific American 253, 68-78 (1985) |
| J.E. Darnell et al: Molecular Cell Biology. Scientific American Books. Freeman, New York, 1990 |
| E.H. McConkey: Human Genetics. The Molecular Revolution. Jones and Bartlett, Boston, 1993. |
Nobel Prizes and laureates
Six prizes were awarded for achievements that have conferred the greatest benefit to humankind. The 12 laureates' work and discoveries range from proteins' structures and machine learning to fighting for a world free of nuclear weapons.
See them all presented here.
