Advanced information
Scientific background:
For the discovery of microRNA and its role in post-transcriptional gene regulation (pdf)
Scientific background
For the discovery of microRNA and its role in post-transcriptional gene regulation
The evolution of multicellular organisms from unicellular ancestors, where each cell-type acquired specialized functions, required increasingly sophisticated mechanisms of gene regulation. Besides transcriptional gene regulation mediated by DNA-binding factors acting on regulatory sequences, other forms of control systems emerged as organisms with increasing complexity evolved. Over hundreds of millions of years, genes encoding tiny non-coding RNA molecules, so-called microRNAs, expanded within genomes of multicellular organisms to exert post-transcriptional control over mRNA stability and protein translation. MicroRNAs and their mode of gene regulation remained completely unknown until the discovery by Victor Ambros and Gary Ruvkun in 1993. The two Nobel laureates investigated mutant C. elegans nematodes with developmental defects caused by alterations at the lin-4 and lin-14 genetic loci. Ambros’s laboratory cloned the lin-4 gene and made the surprising discovery that it did not code for a protein. Instead, it encoded a short 22-nucleotide noncoding RNA. In parallel, Ruvkun’s laboratory determined that lin-4 regulates lin-14 via multiple elements in its 3′ untranslated region (3’UTR). Upon comparing sequence information, they defined partial sequence complementarity between the short non-coding lin-4 RNA and the 3’UTR elements of lin-14. This provided a first glimpse into a conceptually novel type of regulatory RNAs: microRNAs. In 2000, Ruvkun’s lab uncovered the highly conserved let-7 microRNA, leading t the subsequent identification of homologous microRNAs across diverse animal species, including humans. This sparked intense cloning and sequencing efforts to identify microRNAs across the animal kingdom, which led to the finding that microRNAs encompass a large group of regulators that control vast networks of protein-coding genes. The discovery by Ambros and Ruvkun was completely unexpected and unveiled an evolutionarily conserved post-transcriptional regulatory mechanism mediated by microRNAs, with critical roles in animal development and for adult tissue function.
Introduction
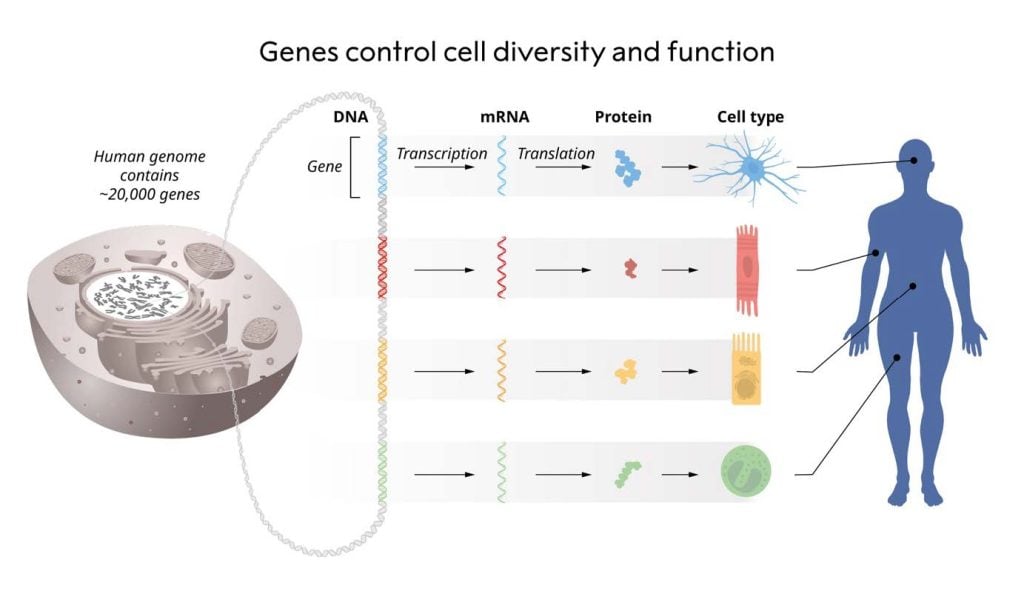
The control of when and where each gene should be transcribed into RNA and translated into protein is a fundamental aspect of life (Figure 1). For instance, insulin is produced in the beta cells of the pancreatic islets, whereas opsins are produced in the retina of the eye. Instructions for precise cell-type specific gene regulation are encoded in the genetic material itself and acted upon by sequence-specific DNA-binding proteins. François Jacob and Jacque Monod were awarded the Nobel Prize in Physiology or Medicine in 1965 for discovering how genes are regulated. The repertoire of DNA-binding transcription factors is well-conserved among unicellular and multicellular eukaryotes (King et al., 2008), whereas additional layers of gene regulation have emerged within multicellular organisms to ensure the correct production of RNA and proteins at any given time in each cell-type.
Eukaryotic model organisms have been invaluable for genetic research, yielding numerous unexpected discoveries. Sydney Brenner introduced the nematode worm Caenorhabditis elegans (C. elegans) over five decades ago. This organism’s short generation time, transparency, and ease of genetic manipulation have facilitated extensive study. Sydney Brenner, John Sulston, and Robert Horvitz used C. elegans to unravel how cell division, differentiation, and cell death are genetically controlled during organ development, discoveries for which they were awarded the 2002 Nobel Prize in Physiology or Medicine.
In the 1970s, mutagenesis screens in C. elegans performed in the Brenner lab uncovered the lin-4 mutant (e912). These worms displayed a striking phenotype: many cell types and morphological structures were entirely absent, and eggs accumulated due to a failure in vulva development (Figure 2) (Horvitz and Sulston, 1980; Chalfie, Horvitz and Sulston, 1981), seemingly from the reiteration of developmental programs for specific cell lineages.
The major disruption of worm development observed in the lin-4 mutant suggested that lin-4 encoded a master regulator of developmental timing. A large number of additional heterochronic mutants exhibiting various temporal developmental defects were characterised, including a second mutant, lin-14, discovered in the Horvitz lab (Ferguson, Sternberg and Horvitz, 1987).
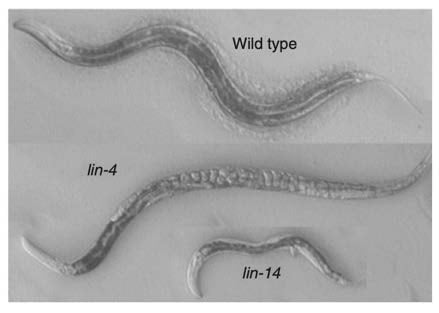
Meanwhile, Victor Ambros joined the Horvitz lab following his PhD with David Baltimore on poliovirus genome structure and replication. As a postdoctoral fellow, Ambros immediately embarked on genetic analyses of heterochronic mutants and identified lin-14 as having developmental timing defects that were opposite to those observed in the lin-4 mutant (Figure 2). In lin-14 mutants, larval programs were completely absent (Ambros and Horvitz, 1984). Notably, Ambros later discovered that lin-4 was a negative regulator of lin-14 (Ambros, 1989).
During this period, Gary Ruvkun had completed his PhD in bacterial genetics under Frederick Ausubel’s supervision. While travelling through Europe, he became intrigued by worm genetics after learning about cell lineage analyses of heterochronic mutants (Chalfie, Horvitz and Sulston, 1981; Ruvkun, Wightman and Ha, 2004). Subsequent discussions with Martin Chalfie and Robert Horvitz further fueled his interest in using C. elegans to investigate these questions. In 1982, Ruvkun started his postdoctoral research jointly between the labs of Walter Gilbert and Robert Horvitz.
Discovery of post-transcriptional gene regulation through microRNAs
In the Horvitz lab, Ambros and Ruvkun started their long pursuit to clone lin-14. At that time, identifying the DNA sequence for a locus defined by genetics was a challenging task. After years of persistent experimentation, they successfully identified the region using a classical restriction fragment length polymorphism approach (Ruvkun et al., 1989). During this period, both Ambros and Ruvkun obtained faculty positions, Ambros at Harvard University and Ruvkun at Massachusetts General Hospital and Harvard Medical School. Committed to their questions, they continued their molecular analyses. Ruvkun demonstrated that lin-14 is a nuclear protein with stage-specific expression in development, high in the L1 stage, and altered in lin-4 and lin-14 mutants (Ruvkun and Giusto, 1989). Interestingly, lin-14 gain-of-function mutants with deletions in the 3’UTR were discovered (Ruvkun and Giusto, 1989; Wightman et al., 1991), leading to prolonged lin-14 protein detection beyond the L1 stage (Arasu, Wightman and Ruvkun, 1991; Wightman et al., 1991). The disruption of 3’UTR elements had no impact on the protein sequence, and Ruvkun, therefore, postulated that a post-transcriptional mechanism acting on mRNA stability, nuclear export, or translation likely mediated the temporal switch in lin-14 (Wightman et al., 1991).
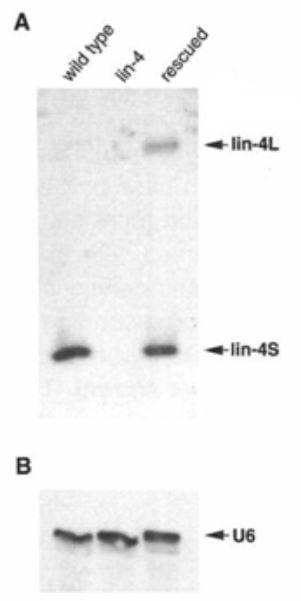
In contrast to the several lin-14 mutants identified, only one mutant in lin-4 had been found (e912). The Ambros lab set out to clone the lin-4 gene guided by restriction fragment length polymorphism and Southern blot probing. “Walking along the chromosome” and iteratively testing smaller genomic fragments for their ability to rescue the mutant lin-4 phenotype, they pinpointed a 693 bp Sal l restriction enzyme fragment. After rounds of open reading frame predictions and clone re-sequencing to rule out errors, they began suspecting that the lin-4 gene might be a noncoding RNA due to its short open reading frame (ORF) sequence. Frameshift mutations introduced into the C. elegans sequence did not affect lin-4 function, confirming this suspicion. In 1991, the lab began probing the lin-4 transcript by Northern blot and RNase protection assay, revealing two short RNA transcripts of 61 and 22 nucleotides (nt) in length (Figure 3).
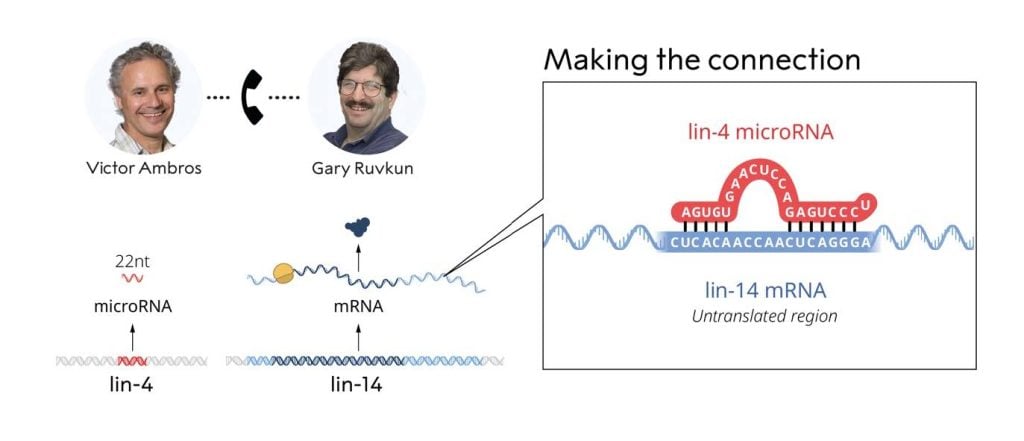
Having independently deduced the sequences of both lin-4 (Ambros lab) and lin-14 (Ruvkun lab), on the evening of June 11, 1992, Ambros and Ruvkun exchanged sequence data for the lin-4 and lin-14 genes. Both noticed conspicuous partial complementarity between the lin-4 noncoding RNA and multiple elements in the lin-14 3’UTR (Figure 4).
Recognizing the significance of their observation, the two labs performed an array of additional experiments demonstrating that the lin-4 microRNA regulates the lin-14 mRNA through base pairing with elements located in the 3’UTR. Their seminal discovery was reported in two papers published back-to-back in Cell in 1993 (Lee, Feinbaum and Ambros, 1993; Wightman, Ha and Ruvkun, 1993).
Ambros’s lab used the C. elegans lin-4 sequence to identify corresponding lin-4 containing clones in other nematodes species (C. briggsae, C. remanei and C. vulgaris). These experiments demonstrated that lin-4 clones from the other nematodes could rescue the lin-4 mutant phenotype in C. elegans. They also screened over 20,000 mutagenized chromosomes to identify a second lin-4 mutant (ma161), which contained a single nucleotide mutation. Notably, this mutation was present within the complementary sequence, further supporting the functional significance of the complementary bases between the lin-4 microRNA and the lin-14 3’UTR elements (Lee, Feinbaum and Ambros, 1993).
Ruvkun’s lab comp ared the amounts of lin-14 protein and RNA in wild-type and lin-14 gain-of-function mutants. Lin-14 protein in the mutants were 4- to 7-fold elevated, with no difference in RNA amounts, demonstrating that lin-14 is regulated at the post-transcriptional level (i.e., after the RNA has been transcribed). Transferring the lin-14 3’UTR into a reporter gene resulted in post-transcriptional regulation of the reporter gene that was similar to lin-14, demonstrating that the heterologous 3’UTR was sufficient to control mRNA translation. Iteratively, smaller fragments of the lin-14 3’UTR were transferred into the reporter until a functional 124 nt long 3’UTR fragment was identified. This 3’UTR region contained several of the partially complementary sequences to lin-4, and additionally this region was conserved in C. briggsae (Wightman, Ha and Ruvkun, 1993).
Computational analysis of the newly discovered lin-4 microRNA against the comprehensive database of nucleotide sequences from all species revealed matching sequences exclusively among other Caenorhabditis nematodes, e.g. C. briggsae. A key question remained: was the presence of microRNA a peculiarity unique to nematodes, or was it conserved with far-reaching functional consequences throughout the animal kingdom?
Discovery of the evolutionarily conserved let-7 microRNA
Following the groundbreaking discovery of lin-4, the first microRNA, seven years passed before a second microRNA gene, let-7, was identified. The Ruvkun lab conducted a genetic screen, focusing on mutants that suppressed the synthetic sterile phenotype of a strain bearing mutations in both the lin-14 and egl-35 loci (Reinhart et al., 2000). Let-7 was found to encode a short 21-nt RNA that exhibited complementarity to 3’UTRs of various heterochronic genes, including lin-14, lin-28, lin-41, lin-42 and daf-12. Loss of let-7 led to the reiteration of larval cell fates in the adult stages. The finding of a second microRNA gene suggested that microRNAs may play a broader role in regulating stage-specific timing of cell lineage formation during development.
The next breakthrough came when the Ruvkun lab found that the let-7 gene, unlike lin-4, was evolutionarily conserved across a wide range of animals. Comparing the let-7 microRNA sequence against nucleotide databases revealed matching sequences in both the fruit fly and human (Pasquinelli et al., 2000). One identified target of let-7 in the nematode was lin-41 (Reinhart et al., 2000), a protein with orthologues in zebrafish and fruit flies. Reassuringly, the 3’UTRs of both the zebrafish and fruit fly lin-41 orthologues showed complementarity to let-7 (Pasquinelli et al., 2000). Furthermore, let-7 microRNA was found across several human tissues indicating its relevance for gene expression in mammalian cells in general.
Similar to nematodes, analysis of fruit fly development demonstrated temporal regulation of the let-7 microRNA, suggesting a conserved role for let-7 among insects, crustaceans, and nematodes (Pasquinelli et al., 2000). Remarkably, temporal let-7 expression was detected even in adult stages of mollusk and annelid species (Pasquinelli et al., 2000), species that do not develop through larval states. Furthermore, vertebrates do not have distinct larval stages but exhibit temporally regulated let-7 expression during development, including strong expression in adult zebrafish. Strikingly, let-7 expression was found to be temporally regulated among bilaterians, i.e. animals with left-right symmetry, and may have evolved after the divergence of these animals from diploblastic species, i.e. those that develop from two primary germ layers instead of three, as do humans and other vertebrates (Figure 5). Discovery of the evolutionarily highly conserved let-7 greatly increased the interest in microRNAs as post-transcriptional regulators of gene expression.
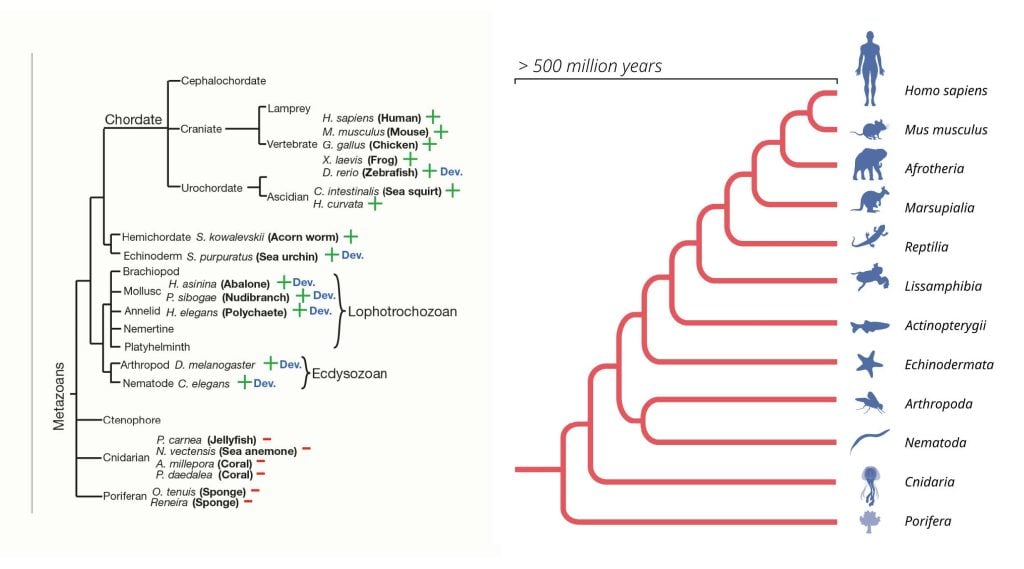
Left: An evolutionary tree of metazoans, highlighting the branches of the tree with a detectable let-7 microRNA
expression (+) or where no let-7 expression was detected (-). Species with similar developmental pattern of let-7 RNA
expression is (no let-7 in early stages; but let-7 expression by adulthood) are indicated by ‘Dev.’. (Pasquinelli et al.,
2000). Right: MicroRNA genes have evolved and expanded within the genomes of multicellular organisms for over 500
million years. © The Nobel Committee for Physiology or Medicine. Ill. Mattias Karlén
Following the discovery of let-7, several research labs sought to identify additional microRNAs in humans and other species via small RNA cloning. The laboratory of Thomas Tuschl cloned novel microRNAs from human and fruit fly tissues (Lagos-Quintana et al., 2001), David Bartel’s lab isolated new microRNAs from the nematode (Lau et al., 2001), as did Ambros lab (Lee and Ambros, 2001). The collective evidence was now compelling: a vast class of regulatory microRNA exists across animals, likely playing important roles in gene regulation. Advances in molecular biology and sequencing technologies have since led to the identification of over a thousand microRNA genes in the human genome. Currently, miRBase, a database for microRNA genes, comprises over 38,000 hairpin precursors and 48,860 mature microRNA gene sequences across 271 organisms (Kozomara, Birgaoanu and Griffiths-Jones, 2019). Even viruses have been found to encode microRNA genes (Pfeffer et al., 2004).
The cloning of additional microRNAs and the availability of whole genome sequences presented increasing opportunities to define the base-pairing rules between microRNAs and 3’UTR regions. Pivotal studies conducted in the laboratories of David Bartel, Christopher Burge and Stephen Cohen (Lewis et al., 2003; Stark et al., 2003; Brennecke et al., 2005; Lewis, Burge and Bartel, 2005) elucidated the overall rules of microRNA target recognition using combined experimental and comparative genomics approaches. These studies showed that microRNAs typically have partial complementarity to the target mRNAs, primarily in the microRNA “seed” region. This work also unveiled that each microRNA likely regulates multiple protein-coding genes, as many 3’UTRs exhibit excessive conservation of sequences complementary to microRNA seed sequences (Brennecke et al., 2005; Lewis, Burge and Bartel, 2005). Interestingly, genes coexpressed with a cell-type or lineage specific microRNA are devoid of target sites for that specific microRNA. In contrast, such microRNA target sites are common in genes expressed in neighboring cells and tissues (Farh et al., 2005; Stark et al., 2005). These observations reinforced the hypothesis that microRNAs have important functions in cell lineage formation and cell-type stability in multicellular organisms.
MicroRNA biogenesis and function
Parallel to the cloning of additional microRNA genes, intense efforts by several research groups were devoted to understanding microRNA biogenesis and mechanisms of action (Bartel, 2004). Strategies for microRNA gene transcription vary. Many microRNA genes are independent transcriptional units, sometimes clustered, while others reside within introns of protein-encoding genes. Canonical primary microRNAs (pri-microRNAs) are transcribed by RNA polymerase II and feature a hairpin-structured sequence. This hairpin serves as a substrate for processing in the nucleus by the microprocessor, a heterotrimeric complex containing Drosha endonuclease, to cleave both strands to produce the precursor microRNA (pre-microRNA), typically 60-70 nucleotides long, first detected in the Ambros lab (Figure 2). Exportin 5 and RAN-GTP facilitate pre-microRNA transport to the cytoplasm. Subsequent processing by Dicer, an endonuclease originally identified in Greg Hannon’s laboratory (Bernstein et al., 2001), forms a microRNA duplex. The effective microRNA strand becomes loaded onto an Argonaute protein-containing silencing complex, whereas the other “passenger” strand is displaced (Schwarz et al., 2003). Once the microRNA strand is loaded into the silencing complex, it can carry out sequence-specific negative regulation of mRNAs via reduced translation and/or mRNA degradation. This regulation involves the adaptor protein TNRC6 and the poly(A)-binding protein PABPC that recruit deadenylase complexes that shorten the mRNA polyA tail, resulting in mRNA degradation and translational inhibition depending on the cellular context, e.g. developmental stage and cell type.
The machinery that processes and executes microRNA function is also used for other RNA-based silencing mechanisms, commonly known as RNA interference (RNAi). These include small interfering RNAs (siRNAs), endogenous piwi-associated RNAs (piRNAs) and repeat-associated small interfering RNA (rasiRNAs). The discovery that double-stranded RNA can induce sequence-dependent gene silencing (Fire et al., 1998), earned Andrew Z. Fire and Craig C. Mello the 2006 Nobel Prize in Physiology or Medicine. Whereas RNAi functions primarily as a defense mechanism against virus infections (in plants and in animals of lower complexity) and against unwanted genomic mobile element activity, microRNAs exert post-transcriptional control over mRNAs throughout development and across adult cell types. To this end, microRNAs have evolved partial complementarity towards their target mRNA sequences to “tune” the respective effects on each mRNA target, whereas e.g. siRNAs are often exogenous with complete complementarity to specific RNA target sequences that become cleaved. In 1999, David Baulcombe showed that post-transcriptional gene silencing in plants involves processing of short RNAs with specificity to target sequences (Hamilton and Baulcombe, 1999), further connecting observations in different fields.
Evolution of microRNAs and their physiological roles
The emergence and expansion of microRNA genes are intimately tied to the evolution of more complex organisms (Figure 5). The number of microRNA genes markedly increased during early bilaterian evolution (Grimson et al., 2008; Wheeler et al., 2009), with their functional roles inferred in the last common bilaterian ancestor prior to protostome and deuterostome divergence (Christodoulou et al., 2010). Since then, hundreds of additional microRNA genes have been gained with the evolution of more specialized cell types and tissues in complex organisms. MicroRNA genes have been identified even in early metazoan sponge, plants, and in two unicellular eukaryote species. Therefore, it is possible that microRNAs may have emerged multiple times during evolution, including the early lineage of multicellular animals around 600 million years ago, or that the ancestor to both plants and animals evolved microRNAs already 1 billion years ago (Moran et al., 2017). Notably, the fact that many evolutionarily old microRNA genes are conserved in later-evolved organisms, and that these genes are rarely lost through evolution, demonstrate their critical roles in gene regulation.
The essential roles of microRNAs in metazoan development and for tissue function have been demonstrated through the ablation of components in the microRNA biogenesis pathway. The loss of Dicer, which processes pre-miRNAs in the cytoplasm, is embryonically lethal in mice and zebrafish (Bernstein et al., 2003; Wienholds et al., 2003). The removal of individual or groups of microRNA genes in fruit flies and mice also causes strong phenotypes (Bartel, 2018). However, the roles of individual microRNA genes can be obscured, likely due to redundant roles of several microRNA genes that share target-defining seed sequences. While the redundancy in the system represents a barrier to studying the function of single microRNA genes, it also demonstrates the robustness of the system and explains why it cannot be easily manipulated by, e.g., viruses.
To highlight the fundamental role of microRNAs, it is important to note that the most evolutionarily conserved microRNA genes, those shared among bilaterian organisms, function early in embryonic development, whereas microRNAs that evolved specifically in mammals function at later stages of embryonic development (DeVeale, Swindlehurst-Chan and Blelloch, 2021). In contrast, species-specific microRNA genes generally play roles in adult cell-types rather that in embryonic development. These patterns are evident from the systematic knockout experiments on microRNA genes of varying evolutionary conservation. The specific regulatory roles of microRNAs during animal development include developmental timing, the formation and stability of cell fates, general physiology, and homeostasis (DeVeale, Swindlehurst-Chan and Blelloch, 2021).
The functions of microRNAs in adult cells and tissues have been elucidated through selective Dicer removal in transgenic mice. Early removal of Dicer1 during B-cell maturation led to a differentiation halt at the pro-B cell stage (Koralov et al., 2008). Dicer1 ablation at embryonic day 15.5 in neurons resulted in the early postnatal death, preceded by microcephaly, reduced dendritic branch elaboration, and increased dendritic spine lengths (Davis et al., 2008). In post-mitotic cerebellar Purkinje cells, Dicer1 loss at two weeks age triggered cerebellar degeneration and ataxia onset (uncoordinated muscle movement) (Schaefer et al., 2007). Similarly, loss of Dicer1 in midbrain dopaminergic neurons led to progressive neuron loss and reduced locomotor activity (Kim et al., 2007). Severe phenotypes have been observed across several other cell types and tissues, demonstrating the critical roles of microRNAs in both developmental processes and adult cell-type functions.
The importance of microRNAs for human development and function becomes apparent through syndromes associated with mutations in specific microRNA genes or components of the biogenesis pathway. The DICER1 syndrome is a rare inherited disorder caused by a mutation in the DICER1 gene, which predisposes individuals to tumors in the kidney, thyroid, ovary, cervix, testicle, brain, eye, and lung. Often, one allele of DICER1 has a germline mutation that renders it nonfunctional, lowering the amounts of functional DICER1 protein in cells. These individuals are vulnerable for additional somatic mutations, and as a consequence, often develop tumors during childhood (Foulkes, Priest and Duchaine, 2014).
The base-pairing portion (i.e., the seed region) of individual microRNA genes is short, making them less likely to be altered by chance mutations. However, there are known mutations in the seed sequence of microRNA genes that are linked to disease. These include mutations in miRNA-96 that are associated with progressive hearing loss (Mencía et al., 2009; Soldà et al., mutations in miRNA-184 causing EDICT syndrome, a rare eye disease with iris hypoplasia, endothelial dystrophy, and congenital cataract (Hughes et al., 2011; Iliff, Riazuddin and Gottsch, 2012; Lechner et al., 2013), and mutations in miRNA-140-5p resulting in a congenital skeletal disorder (Grigelioniene et al., 2019). Advances are being made in developing microRNA-based diagnostics and therapeutics for diseases, such as metabolic disorders, cardiovascular disease, neurodegenerative conditions, and cancer.
Summary
Thanks to the seminal discovery by Ambros and Ruvkun, and the many colleagues who built on their findings, a new dimension to gene regulation has been revealed. Whereas proteins in the nucleus regulate RNA transcription and splicing, microRNAs control the translation and degradation of mRNA in the cytoplasm. This unexpected layer of post-transcriptional gene regulation has critical importance throughout animal development and in adult cell types and is essential for complex multicellular life.
Rickard Sandberg, PhD, Professor at Karolinska Institutet, (Rickard.Sandberg@ki.se), Member of the Nobel Committee
Key references
Rosalind C. Lee, Rhonda L. Feinbaum and Victor Ambros (1993) “The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14”. Cell, 75(5), pp. 843–854.
Bruce Wightman, Ilho Ha, and Gary Ruvkun (1993) “Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans”. Cell, 75(5), pp. 855–862.
Amy E. Pasquinelli, Brenda J. Reinhart, Frank Slack, Mark Q. Martindale, Mitzi I. Kurodak, Betsy Maller, David C. Hayward, Eldon E. Ball, Bernard Degnan, Peter Müller, Jürg Spring, Ashok Srinivasan, Mark Fishman, John Finnerty, Joseph Corbo, Michael Levine, Patrick Leahy, Eric Davidson & Gary Ruvkun (2000) “Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA”. Nature, 408(6808), pp. 86–89.
*these authors contributed equally to this work.
The Nobel Assembly, consisting of 50 professors at Karolinska Institutet, awards the Nobel Prize in Physiology or Medicine. Its Nobel Committee evaluates the nominations. Since 1901 the Nobel Prize has been awarded to scientists who have made the most important discoveries for the benefit of humankind.
Nobel Prize® is the registered trademark of the Nobel Foundation
References
Ambros, V. (1989) ‘A hierarchy of regulatory gene controls a larva-to-adult developmental switch in C. elegans’, Cell 57(1), pp. 49–57. Available at: https://doi.org/10.1016/0092-8674(89)90171- 2.
Ambros, V. (2008) ‘The evolution of our thinking about microRNAs’, Nature Medicine, 14(10), pp. 1036–1040. Available a https://doi.org/10.1038/nm1008-1036.
Ambros, V. and Horvitz, H.R. (1984) ‘Hetero’chroni mutants of the nematode Caenorhabditis elegans’, Science (New York, N.Y.), 226(4673), pp. 409–416. Available at: https://doi.org/10.1126/science.64948 91.
Arasu, P., Wightman, B. and Ruvkun, G. (1991) ‘Temporal regulation of lin-14 by the antagonistic action of two other heterochronic genes, lin-4 and lin-28’, Genes & Development, 5(10), pp. 1825–1833. Available at: https://doi.org/10.1101/gad.5.10.1825.
Bartel, D.P. (2004) ‘MicroRNAs: genomics, biogenes is, mechanism, and function’, Cell, 116(2), pp. 281–297. Available a https://doi.org/10.1016/s0092-8674(04)00045-5.
Bartel, D.P. (2018) ‘Metazoan MicroRNAs’, Cell, 173(1), pp. 20–51. Available at: https://doi.org/10.1016/j.cell.2018.03.006 .
Bernstein, E. et al. (2001) ‘Role for bidentate ribonuclease in the initiation step of RNA interference’, Nature, 409(6818), pp. 363–366. Available at: https://doi.org/10.1038/35053110.
Bernstein, E. et al. (2003) ‘Dicer is essential for mouse development’, Nature Genetics, 35(3), pp. 215–217. Available at: https://doi.org/10.1038/ng1253.
Brennecke, J. et al. (2005) ‘Principles o microRNA-target recognition’, PLoS biology, 3(3), p. e85. Available at: https://doi.org/10.1371/journal.pbio.0030085.
Chalfie, M., Horvitz, H.R. and Sulston, J.E. (1981) ‘Mutations that lead to reiterations in the cell lineages of C. elegans’, Cell, 24(1), pp. 59–69. Available at: https://doi.org/10.1016/0092-8674(81)90501-8.
Christodoulou, F. et al. (2010) ‘Ancient animal microRNAs and the evolution of tissue identity’, Nature, 463(7284), pp. 1084–1088. Available at: https://doi.org/10.1038/nature08744.
Davis, T.H. et al. (2008) ‘Conditional Loss of Dicer Disrupts Cellular and Tissue Morphogenesis in the Cortex and Hippocampus’, The Journal of Neuroscience, 28(17), pp. 4322–4330. Available at: https://doi.org/10.1523/JNEUROSCI.4815-07.2008.
DeVeale, B., Swindlehurst-Chan, J. and Blelloch, R. (2021) ‘The roles of microRNAs in mouse development’, Nature Reviews. Genetics, 22(5), pp. 307–323. Available at: https://doi.org/10.1038/s41576-020-00309-5.
Farh, K.K.-H. et al. (2005) ‘The widespread impact of mammalian MicroRNAs on mRNA repression and evolution’, Science (New York, N.Y.), 310(5755), pp. 1817–1821. Available at: https://doi.org/10.1126/science.1121158.
Ferguson, E.L., Sternberg, P.W. and Horvitz, H.R. (1987) ‘A genetic pathway for the specification of the vulval cell lineages of Caenorhabditis elegans’, Nature, 326(6110), pp. 259–267. Available at: https://doi.org/10.1038/326259a0.
Fire, A. et al. (1998) ‘Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans’, Nature, 391(6669), pp. 806–811. Available at: https://doi.org/10.1038/35888.
Foulkes, W.D., Priest, J.R. and Duchaine, T.F. (2014) ‘DICER1: mutations, microRNAs and mechanisms’, Nature Reviews. Cancer, 14(10), pp. 662–672. Available at: https://doi.org/10.1038/nrc3802.
Grigelioniene, G. et al. (2019) ‘Gain-of-function mutation of microRNA-140 in human skeletal dysplasia’, Nature Medicine, 25(4), pp. 583–590. Available at: https://doi.org/10.1038/s41591-019-0353-2.
Grimson, A. et al. (2008) ‘Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals’, Nature, 455(7217), pp. 1193–1197. Available at: https://doi.org/10.1038/nature07415.
Hamilton, A.J. and Baulcombe, D.C. (1999) ‘A species of small antisense RNA in posttranscriptional gene silencing in plants’, Science (New York, N.Y.), 286(5441), pp. 950–952. Available at: https://doi.org/10.1126/science.286.5441.950.
Horvitz, H.R. and Sulston, J.E. (1980) ‘Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans’, Genetics, 96(2), pp. 435–454. Available at: https://doi.org/10.1093/genetics/96.2.435.
Hughes, A.E. et al. (2011) ‘Mutation altering the miR-184 seed region causes familial keratoconus with cataract’, American Journal of Human Genetics, 89(5), pp. 628–633. Available at: https://doi.org/10.1016/j.ajhg.2011.09.014.
Iliff, B.W., Riazuddin, S.A. and Gottsch, J.D. (2012) ‘A single-base substitution in the seed region of miR-184 causes EDICT syndrome’, Investigative Ophthalmology & Visual Science, 53(1), pp. 348–353. Available at: https://doi.org/10.1167/iovs.11-8783.
Kim, J. et al. (2007) ‘A MicroRNA feedback circuit in midbrain dopamine neurons’, Science (New York, N.Y.), 317(5842), pp. 1220–1224. Available at: https://doi.org/10.1126/science.1140481.
King, N. et al. (2008) ‘The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans’, Nature, 451(7180), pp. 783–788. Available at: https://doi.org/10.1038/nature06617.
Koralov, S.B. et al. (2008) ‘Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage’, Cell, 132(5), pp. 860–874. Available at: https://doi.org/10.1016/j.cell.2008.02.020.
Kozomara, A., Birgaoanu, M. and Griffiths-Jones, S. (2019) ‘miRBase: from microRNA sequences to function’, Nucleic Acids Research, 47(D1), pp. D155–D162. Available at: https://doi.org/10.1093/nar/gky1141.
Lagos-Quintana, M. et al. (2001) ‘Identification of novel genes coding for small expressed RNAs’, Science (New York, N.Y.), 294(5543), pp. 853–858. Available at: https://doi.org/10.1126/science.1064921.
Lau, N.C. et al. (2001) ‘An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans’, Science (New York, N.Y.), 294(5543), pp. 858–862. Available at: https://doi.org/10.1126/science.1065062.
Lechner, J. et al. (2013) ‘Mutational analysis of MIR184 in sporadic keratoconus and myopia’, Investigative Ophthalmology & Visual Science, 54(8), pp. 5266–5272. Available at: https://doi.org/10.1167/iovs.13-12035.
Lee, R.C. and Ambros, V. (2001) ‘An extensive class of small RNAs in Caenorhabditis elegans’, Science (New York, N.Y.), 294(5543), pp. 862–864. Available at: https://doi.org/10.1126/science.1065329.
Lee, R.C., Feinbaum, R.L. and Ambros, V. (1993) ‘The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14’, Cell, 75(5), pp. 843–854. Available at: https://doi.org/10.1016/0092-8674(93)90529-y.
Lewis, B.P. et al. (2003) ‘Prediction of mammalian microRNA targets’, Cell, 115(7), pp. 787–798. Available at: https://doi.org/10.1016/s0092-8674(03)01018-3.
Lewis, B.P., Burge, C.B. and Bartel, D.P. (2005) ‘Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets’, Cell, 120(1), pp. 15–20. Available at: https://doi.org/10.1016/j.cell.2004.12.035.
Mencía, A. et al. (2009) ‘Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss’, Nature Genetics, 41(5), pp. 609–613. Available at: https://doi.org/10.1038/ng.355.
Moran, Y. et al. (2017) ‘The evolutionary origin of plant and animal microRNAs’, Nature Ecology & Evolution, 1(3), pp. 1–8. Available at: https://doi.org/10.1038/s41559-016-0027.
Pasquinelli, A.E. et al. (2000) ‘Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA’, Nature, 408(6808), pp. 86–89. Available at: https://doi.org/10.1038/35040556.
Pfeffer, S. et al. (2004) ‘Identification of virus-encoded microRNAs’, Science (New York, N.Y.), 304(5671), pp. 734–736. Available at: https://doi.org/10.1126/science.1096781.
Reinhart, B.J. et al. (2000) ‘The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans’, Nature, 403(6772), pp. 901–906. Available at: https://doi.org/10.1038/35002607.
Ruvkun, G. et al. (1989) ‘Molecular genetics of the Caenorhabditis elegans heterochronic gene lin-14’, Genetics, 121(3), pp. 501–516. Available at: https://doi.org/10.1093/genetics/121.3.501.
Ruvkun, G. and Giusto, J. (1989) ‘The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch’, Nature, 338(6213), pp. 313–319. Available at: https://doi.org/10.1038/338313a0.
Ruvkun, G., Wightman, B. and Ha, I. (2004) ‘The 20 years it took to recognize the importance of tiny RNAs’, Cell, 116(2 Suppl), pp. S93-96, 2 p following S96. Available at: https://doi.org/10.1016/s0092-8674(04)00034-0.
Schaefer, A. et al. (2007) ‘Cerebellar neurodegeneration in the absence of microRNAs’, Journal of Experimental Medicine, 204(7), pp. 1553–1558. Available at: https://doi.org/10.1084/jem.20070823.
Schwarz, D.S. et al. (2003) ‘Asymmetry in the assembly of the RNAi enzyme complex’, Cell, 115(2), pp. 199–208. Available at: https://doi.org/10.1016/s0092-8674(03)00759-1.
Soldà, G. et al. (2012) ‘A novel mutation within the MIR96 gene causes non-syndromic inherited hearing loss in an Italian family by altering pre-miRNA processing’, Human Molecular Genetics, 21(3), pp. 577–585. Available at: https://doi.org/10.1093/hmg/ddr493.
Stark, A. et al. (2003) ‘Identification of Drosophila MicroRNA targets’, PLoS biology, 1(3), p. E60. Available at: https://doi.org/10.1371/journal.pbio.0000060.
Stark, A. et al. (2005) ‘Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3’UTR evolution’, Cell, 123(6), pp. 1133–1146. Available at: https://doi.org/10.1016/j.cell.2005.11.023.
Wheeler, B.M. et al. (2009) ‘The deep evolution of metazoan microRNAs’, Evolution & Development, 11(1), pp. 50–68. Available at: https://doi.org/10.1111/j.1525-142X.2008.00302.x.
Wienholds, E. et al. (2003) ‘The microRNA-producing enzyme Dicer1 is essential for zebrafish development’, Nature Genetics, 35(3), pp. 217–218. Available at: https://doi.org/10.1038/ng1251.
Wightman, B. et al. (1991) ‘Negative regulatory sequences in the lin-14 3’-untranslated region are necessary to generate a temporal switch during Caenorhabditis elegans development’, Genes & Development, 5(10), pp. 1813–1824. Available at: https://doi.org/10.1101/gad.5.10.1813.
Wightman, B., Ha, I. and Ruvkun, G. (1993) ‘Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans’, Cell, 75(5), pp. 855–862. Available at: https://doi.org/10.1016/0092-8674(93)90530-4.
Nobel Prizes and laureates
Six prizes were awarded for achievements that have conferred the greatest benefit to humankind. The 12 laureates' work and discoveries range from proteins' structures and machine learning to fighting for a world free of nuclear weapons.
See them all presented here.
